Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates
- PMID: 11679338
- PMCID: PMC93283
- DOI: 10.1128/AEM.67.11.5143-5153.2001
Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates
Abstract
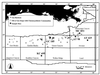
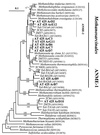
Although there is significant interest in the potential interactions of microbes with gas hydrate, no direct physical association between them has been demonstrated. We examined several intact samples of naturally occurring gas hydrate from the Gulf of Mexico for evidence of microbes. All samples were collected from anaerobic hemipelagic mud within the gas hydrate stability zone, at water depths in the ca. 540- to 2,000-m range. The delta(13)C of hydrate-bound methane varied from -45.1 per thousand Peedee belemnite (PDB) to -74.7 per thousand PDB, reflecting different gas origins. Stable isotope composition data indicated microbial consumption of methane or propane in some of the samples. Evidence of the presence of microbes was initially determined by 4,6-diamidino 2-phenylindole dihydrochloride (DAPI) total direct counts of hydrate-associated sediments (mean = 1.5 x 10(9) cells g(-1)) and gas hydrate (mean = 1.0 x 10(6) cells ml(-1)). Small-subunit rRNA phylogenetic characterization was performed to assess the composition of the microbial community in one gas hydrate sample (AT425) that had no detectable associated sediment and showed evidence of microbial methane consumption. Bacteria were moderately diverse within AT425 and were dominated by gene sequences related to several groups of Proteobacteria, as well as Actinobacteria and low-G + C Firmicutes. In contrast, there was low diversity of Archaea, nearly all of which were related to methanogenic Archaea, with the majority specifically related to Methanosaeta spp. The results of this study suggest that there is a direct association between microbes and gas hydrate, a finding that may have significance for hydrocarbon flux into the Gulf of Mexico and for life in extreme environments.
Figures

References
-
- Aharon P, Scharez H P, Roberts H H. Radiometric dating of hydrocarbon seeps in the Gulf of Mexico. GSA Bull. 1997;109:568–579.
-
- Balows A, Truber H G, Dworkin M, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992.
-
- Bidle K A, Kastner M, Bartlett D H. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia Margin (ODP Site 892B) FEMS Microbiol Ecol. 1999;177:101–108. - PubMed
-
- Blaut M. Metabolism of methanogens. Antonie Leeuwenhoek. 1994;66:187–208. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

