Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: implications for vaccine and antiretroviral control strategies
- PMID: 12551997
- PMCID: PMC141090
- DOI: 10.1128/jvi.77.4.2587-2599.2003
Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: implications for vaccine and antiretroviral control strategies
Abstract
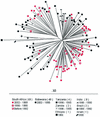
The KwaZulu-Natal region of South Africa is experiencing an explosive outbreak of human immunodeficiency virus type 1 (HIV-1) subtype C infections. Understanding the genetic diversity of C viruses and the biological consequences of this diversity is important for the design of effective control strategies. We analyzed the protease gene, the first 935 nucleotides of reverse transcriptase, and the C2V5 envelope region of a representative set of 72 treatment-naïve patients from KwaZulu-Natal and correlated the results with amino acid signature and resistance patterns. Phylogenetic analysis revealed multiple clusters or "lineages" of HIV-1 subtype C that segregated with other C viruses from southern Africa. The same pattern was observed for both black and Indian subgroups and for retrospective specimens collected prior to 1990, indicating that multiple sublineages of HIV-1 C have been present in KwaZulu-Natal since the early stages of the epidemic. With the exception of three nonnucleoside reverse transcriptase inhibitor mutations, no primary resistance mutations were identified. Numerous accessory polymorphisms were present in the protease, but none were located at drug-binding or active sites of the enzyme. One frequent polymorphism, I93L, was located near the protease/reverse transcriptase cleavage site. In the envelope, disruption of the glycosylation motif at the beginning of V3 was associated with the presence of an extra protein kinase C phosphorylation site at codon 11. Many polymorphisms were embedded within cytotoxic T lymphocyte or overlapping cytotoxic T-lymphocyte/T-helper epitopes, as defined for subtype B. This work forms a baseline for future studies aimed at understanding the impact of genetic diversity on vaccine efficacy and on natural susceptibility to antiretroviral drugs.
Figures



References
-
- Akaike, H. 1997. A new look at statistical model identification. IEEE Trans. Automatic Control 19:716-723.
-
- Biggar, R. J., Janes, M., Pilon, R., Roy, R., Broadhead, R., Kumwenda, N., Taha, T. E., and S. Cassol. 2002. Human immunodeficiency virus type 1 infection in twin pairs infected at birth. J. Infect. Dis. 186:281-285. - PubMed
-
- Cassol, S., B. G. Weniger, G. Babu, et al. 1996. Detection of HIV-1 env subtypes A, B, C, and E in Asia with dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res. Hum. Retrovir. 12:1435-1441. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

