Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum
- PMID: 14711690
- PMCID: PMC321288
- DOI: 10.1128/AEM.70.1.581-587.2004
Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum
Abstract
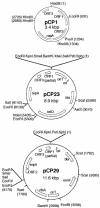
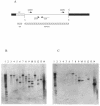
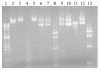
Flavobacterium psychrophilum, a member of the Cytophaga-Flavobacterium-Bacteroides group, is an important pathogen of salmonid fish. Previous attempts to develop genetic techniques for this fastidious, psychrotrophic bacterium have met with failure. Here we describe the development of techniques for the genetic manipulation of F. psychrophilum and the identification of plasmids, selectable markers, a reporter system, and a transposon that function in several isolates of this fish pathogen. The antibiotic resistance genes ermF, cfxA, and tetQ function in F. psychrophilum. Cloning vectors based on the F. psychrophilum cryptic plasmid pCP1 which carried these selectable markers were introduced by conjugation from E. coli, resulting in antibiotic-resistant colonies of F. psychrophilum. Conjugative transfer of DNA into F. psychrophilum was strain dependent. Efficient transfer was observed for two of the seven strains tested (THC02-90 and THC04-90). E. coli lacZY functioned in F. psychrophilum when expressed from a pCP1 promoter, allowing its development as a reporter for studies of gene expression. Plasmids isolated from F. psychrophilum were efficiently introduced into F. psychrophilum by electroporation, but plasmids isolated from E. coli were not suitable for transfer by this route, suggesting the presence of a restriction barrier. DNA isolated from F. psychrophilum was resistant to digestion by Sau3AI and BamHI, indicating that a Sau3AI-like restriction modification system may constitute part of this barrier. Tn4351 was introduced into F. psychrophilum from E. coli and transposed with apparent randomness, resulting in erythromycin-resistant colonies. The techniques developed in this study allow for genetic manipulation and analysis of this important fish pathogen.
Figures

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bertolini, J. M., H. Wakabayashi, V. G. Watral, M. J. Whipple, and J. S. Rohovec. 1994. Electrophoretic detection of proteases from selected strains of Flexibacter psychrophilus and assessment of their variability. J. Aquat. Anim. Health 6:224-233.
-
- Chakroun, C., F. Grimont, M. C. Urdaci, and J.-F. Bernardet. 1998. Fingerprinting of Flavobacterium psychrophilum isolates by ribotyping and plasmid profiling. Dis. Aquat. Org. 33:167-177. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

