Horizontal gene transfer from flowering plants to Gnetum
- PMID: 12963817
- PMCID: PMC196887
- DOI: 10.1073/pnas.1833775100
Horizontal gene transfer from flowering plants to Gnetum
Abstract
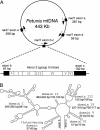
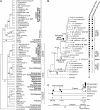
Although horizontal gene transfer is well documented in microbial genomes, no case has been reported in higher plants. We discovered horizontal transfer of the mitochondrial nad1 intron 2 and adjacent exons b and c from an asterid to Gnetum (Gnetales, gymnosperms). Gnetum has two copies of intron 2, a group II intron, that differ in their exons, nucleotide composition, domain lengths, and structural characteristics. One of the copies, limited to an Asian clade of Gnetum, is almost identical to the homologous locus in angiosperms, and partial sequences of its exons b and c show characteristic substitutions unique to angiosperms. Analyses of 70 seed plant nad1 exons b and c and intron 2 sequences, including representatives of all angiosperm clades, support that this copy originated from a euasterid and was horizontally transferred to Gnetum. Molecular clock dating, using calibrations provided by gnetalean macrofossils, suggests an age of 5 to 2 million years for the Asian clade that received the horizontal transfer.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

