Adeno-associated viruses undergo substantial evolution in primates during natural infections
- PMID: 12716974
- PMCID: PMC156329
- DOI: 10.1073/pnas.0937739100
Adeno-associated viruses undergo substantial evolution in primates during natural infections
Abstract
Adeno-associated viruses (AAVs) are single-stranded DNA viruses that are endemic in human populations without known clinical sequelae and are being evaluated as vectors for human gene therapy. To better understand the biology of this virus, we examined a number of nonhuman primate species for the presence of previously uncharacterized AAVs and characterized their structure and distribution. AAV genomes were widely disseminated throughout multiple tissues of a variety of nonhuman primate species. Surprising diversity of sequence, primarily localized to hypervariable regions of the capsid protein, was detected. This diversity of sequence is caused, in part, by homologous recombination of co-infecting parental viruses that modify the serologic reactivity and tropism of the virus. This is an example of rapid molecular evolution of a DNA virus in a way that was formerly thought to be restricted to RNA viruses.
Figures

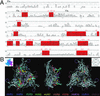
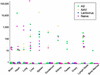


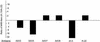
References
-
- Muzyczka N, Berns K I. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 2. Williams & Wilkins, Philadelphia: Lippincott; 2001. pp. 2327–2359.
-
- Kotin R M. Hum Gene Ther. 1994;5:793–801. - PubMed
-
- Atchison R W, Casto B C, Hammon W M. Science. 1965;149:754–756. - PubMed
-
- Mayor H D, Melnick J L. Nature. 1966;210:331–332. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

