Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing
- PMID: 12782724
- PMCID: PMC156367
- DOI: 10.1105/tpc.009548
Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing
Abstract
In Arabidopsis thaliana, cis-regulatory sequences of the floral homeotic gene AGAMOUS (AG) are located in the second intron. This 3-kb intron contains binding sites for two direct activators of AG, LEAFY (LFY) and WUSCHEL (WUS), along with other putative regulatory elements. We have used phylogenetic footprinting and the related technique of phylogenetic shadowing to identify putative cis-regulatory elements in this intron. Among 29 Brassicaceae species, several other motifs, but not the LFY and WUS binding sites identified previously, are largely invariant. Using reporter gene analyses, we tested six of these motifs and found that they are all functionally important for the activity of AG regulatory sequences in A. thaliana. Although there is little obvious sequence similarity outside the Brassicaceae, the intron from cucumber AG has at least partial activity in A. thaliana. Our studies underscore the value of the comparative approach as a tool that complements gene-by-gene promoter dissection but also demonstrate that sequence-based studies alone are insufficient for a complete identification of cis-regulatory sites.
Figures

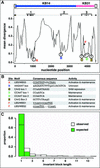





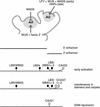
References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Bergman, C.M., and Kreitman, M. (2001). Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11, 1335–1345. - PubMed
-
- Berman, B.P., Nibu, Y., Pfeiffer, B.D., Tomancak, P., Celniker, S.E., Levine, M., Rubin, G.M., and Eisen, M.B. (2002). Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99, 757–762. - PMC - PubMed
-
- Blázquez, M.A., Soowal, L., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835–3844. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

