Comparison of TFII-I gene family members deleted in Williams-Beuren syndrome
- PMID: 15388857
- PMCID: PMC2286546
- DOI: 10.1110/ps.04747604
Comparison of TFII-I gene family members deleted in Williams-Beuren syndrome
Abstract
Williams-Beuren syndrome (WBS) is a neurological disorder resulting from a microdeletion, typically 1.5 megabases in size, at 7q11.23. Atypical patients implicate genes at the telomeric end of this multigene deletion as the main candidates for the pathology of WBS in particular the unequal cognitive profile associated with the condition. We recently identified a gene (GTF2IRD2) that shares homology with other members of a unique family of transcription factors (TFII-I family), which reside in the critical telomeric region. Using bioinformatics tools this study focuses on the detailed assessment of this gene family, concentrating on their characteristic structural components such as the leucine zipper (LZ) and I-repeat elements, in an attempt to identify features that could aid functional predictions. Phylogenetic analysis identified distinct I-repeat clades shared between family members. Linking functional data to one such clade has implicated them in DNA binding. The identification of PEST, synergy control motifs, and sumoylation sites common to all family members suggest a shared mechanism regulating the stability and transcriptional activity of these factors. In addition, the identification/isolation of short truncated isoforms for each TFII-I family member implies a mode of self-regulation. The exceptionally high identity shared between GTF2I and GTF2IRD2, suggests that heterodimers as well as homodimers are possible, and indicates overlapping functions between their respective short isoforms. Such cross-reactivity between GTF2I and GTF2IRD2 short isoforms might have been the evolutionary driving force for the 7q11.23 chromosomal rearrangement not present in the syntenic region in mice.
Figures
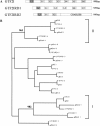
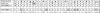



References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Baumer, A., Dutly, F., Balmer, D., Riegel, M., Tukel, T., Krajewska-Walasek, M., and Schinzel, A. 1998. High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum. Mol. Genet. 7 887–894. - PubMed
-
- Bayarsaihan, D., Dunai, J., Greally, J.M., Kawasaki, K., Sumiyama, K., Enkhmandakh, B., Shimizu, N., and Ruddle, F.H. 2002. Genomic organization of the genes Gtf2ird1, Gtf2i and Ncf1 at the mouse chromosome 5 region syntenic to the human chromosome 7q11.23 Williams syndrome critical region. Genomics 79 137–143. - PubMed
-
- Bellugi, U., Lichtenberger, L., Jones, W., and Lai, Z. 2000. The neurocognitive profile of Williams syndrome: A complex pattern of strengths and weaknesses. J. Cogn. Neurosci. 12 (Suppl. 1) 7–29. - PubMed
-
- Benezra, R., Davis, R.L., Lockshun, D., Turner, D.L., and Weintraub, H. 1990. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell 61 49–59. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

