Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development
- PMID: 14526020
- PMCID: PMC225022
- DOI: 10.1128/JB.185.20.6083-6094.2003
Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development
Abstract
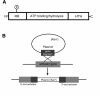
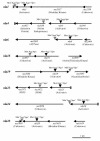
The multicellular developmental cycle of Myxococcus xanthus requires large-scale changes in gene transcription, and recent findings indicate that NtrC-like activators play a prominent role in regulating these changes. In this study, we made insertions in 28 uncharacterized ntrC-like activator (nla) genes and found that eight of these insertions cause developmental defects. Hence, these results are consistent with the idea that M. xanthus uses a series of different NtrC-like activators during fruiting body development. Four of the eight developmental mutants we identified have motility defects. The nla1, nla19, and nla23 mutants show S-motility defects, while the nla24 mutant shows defects in both S-motility and A-motility. During development, aggregation of the nla1, nla19, and nla23 mutants is delayed slightly and the nla24 mutant shows no signs of aggregation or sporulation. The nla4, nla6, nla18, and nla28 mutants have no appreciable loss in motility, but they fail to aggregate and to sporulate normally. The nla18 mutant belongs to a special class of developmental mutants whose defects can be rescued when they are codeveloped with wild-type cells, suggesting that nla18 fails to produce a cell-cell signal required for development. The three remaining activator mutants, nla4, nla6, and nla28, appear to have complex developmental phenotypes that include deficiencies in cell-cell developmental signals.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

