Denaturing gradient gel electrophoretic analysis of ammonia-oxidizing bacterial community structure in the lower Seine River: impact of Paris wastewater effluents
- PMID: 15528539
- PMCID: PMC525213
- DOI: 10.1128/AEM.70.11.6726-6737.2004
Denaturing gradient gel electrophoretic analysis of ammonia-oxidizing bacterial community structure in the lower Seine River: impact of Paris wastewater effluents
Abstract
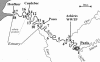
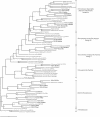
The Seine River is strongly affected by the effluents from the Acheres wastewater treatment plant (WWTP) downstream of the city of Paris. We have shown that the effluents introduce large amounts of ammonia and inoculate the receiving medium with nitrifying bacteria. The aim of the present study was to investigate the diversity of the ammonia-oxidizing bacterial population by identifying autochthonous bacteria from upstream and/or allochthonous ammonia-oxidizing bacteria from the WWTP effluents. Measurements of potential nitrifying activity, competitive PCR, and denaturing gradient gel electrophoresis (DGGE) of 16S ribosomal DNA fragments specific to ammonia-oxidizing bacteria (AOB) were used to explore the succession and shifts of the ammonia-oxidizing community in the lower Seine River and to analyze the temporal and spatial functioning of the system at several different sampling dates. A major revelation was the stability of the patterns. The CTO primers used in this study (G. A. Kowalchuk, J. R. Stephen, W. D. Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp, Appl. Environ. Microbiol. 63:1489-1497, 1997) were shown not to be completely specific to AOB of the beta subclass of Proteobacteria. We further demonstrated that when DGGE patterns are interpreted, all the different bands must be sequenced, as one major DGGE band proved to be affiliated with a group of non-AOB in the beta subclass of Proteobacteria. The majority of AOB (75 to 90%) present in the lower Seine river downstream of the effluent output belong to lineage 6a, represented by Nitrosomonas oligotropha- and Nitrosomonas ureae-like bacteria. This dominant lineage was represented by three bands on the DGGE gel. The major lineage-6a AOB species, introduced by the WWTP effluents, survived and might have grown in the receiving medium far downstream, in the estuary; it represented about 40% of the whole AOB population. The other two species belonging to lineage 6a seem to be autochthonous bacteria. One of them developed a few kilometers downstream of the WWTP effluent input in an ammonia-enriched environment, and the other appeared in the freshwater part of the estuary and was apparently more adapted to estuarine conditions, i.e., an increase in the amount of suspended matter, a low ammonia concentration, and high turnover of organic matter. The rest of the AOB population was represented in equal proportions by Nitrosospira- and Nitrosococcus mobilis-like species.
Figures




References
-
- Aakra, A., J. B. Utaker, and I. F. Nes. 2001. Comparative phylogeny of the ammonia monooxygenase subunit A and 16S rRNA genes of ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 205:237-242. - PubMed
-
- Avrahami, S., W. Liesack, and R. Conrad. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 5:691-705. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

