A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma
- PMID: 15753289
- PMCID: PMC554800
- DOI: 10.1073/pnas.0407918102
A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma
Abstract
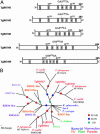
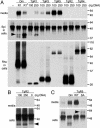
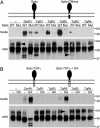
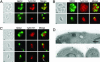
Apicomplexan parasites cause serious human and animal diseases, the treatment of which requires identification of new therapeutic targets. Host-cell invasion culminates in the essential cleavage of parasite adhesins, and although the cleavage site for several adhesins maps within their transmembrane domains, the protease responsible for this processing has not been discovered. We have identified, cloned, and characterized the five nonmitochondrial rhomboid intramembrane proteases encoded in the recently completed genome of Toxoplasma gondii. Four T. gondii rhomboids (TgROMs) were active proteases with similar substrate specificity. TgROM1, TgROM4, and TgROM5 were expressed in the tachyzoite stage responsible for the disease, whereas TgROM2 and TgROM3 were expressed in the oocyst stage involved in transmission. Although both TgROM5 and TgROM4 localized to the cell surface in tachyzoites, TgROM5 was primarily at the posterior of the parasite, whereas adhesins were sequestered in internal micronemes. Upon microneme secretion, as occurs during invasion, the MIC2 adhesin was secreted to the apical end and translocated to the posterior, the site of cleavage, where it colocalized only with TgROM5. Moreover, only TgROM5 was able to cleave MIC adhesins in a cell-based assay, indicating that it likely provides the key protease activity necessary for invasion. T. gondii rhomboids have clear homologues in other apicomplexans including malaria; thus, our findings provide a model for studying invasion by this deadly pathogen and offer a target for therapeutic intervention.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

