Laboratory-based surveillance and molecular epidemiology of influenza virus in Taiwan
- PMID: 15814980
- PMCID: PMC1081360
- DOI: 10.1128/JCM.43.4.1651-1661.2005
Laboratory-based surveillance and molecular epidemiology of influenza virus in Taiwan
Abstract
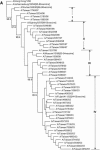
A laboratory-based surveillance network of 11 clinical virological laboratories for influenza viruses was established in Taiwan under the coordination of the Center for Disease Control and Prevention (CDC), Taiwan. From October 2000 to March 2004, 3,244 influenza viruses were isolated, including 1,969 influenza A and 1,275 influenza B viruses. The influenza infections usually occurred frequently in winter in the northern hemisphere. However, the influenza seasonality in Taiwan was not clear during the four seasons under investigation. For example, the influenza A viruses peaked during the winters of 2001, 2002, and 2003. However, some isolated peaks were also found in the summer and fall (June to November) of 2001 and 2002. An unusual peak of influenza B also occurred in the summer of 2002 (June to August). Phylogenetic analysis shows that influenza A isolates from the same year were often grouped together. However, influenza B isolates from the year 2002 clustered into different groups, and the data indicate that both B/Victoria/2/87-like and B/Yamagata/16/88-like lineages of influenza B viruses were cocirculating. Sequence comparison of epidemic strains versus vaccine strains shows that many vaccine-like Taiwanese strains were circulating at least 2 years before the vaccine strains were introduced. No clear seasonality of influenza reports in Taiwan occurred in contrast to other more continental regions.
Figures






References
-
- Barr, I. G., N. Komadina, A. Hurt, R. Shaw, C. Durrant, P. Iannello, C. Tomasov, H. Sjogren, and A. W. Hampson. 2003. Reassortants in recent human influenza A and B isolates from South East Asia and Oceania. Virus Res. 98:35-44. - PubMed
-
- Besselaar, T. G., L. Botha, J. M. McAnerney, and B. D. Schoub. 2004. Phylogenetic studies of influenza B viruses isolated in southern Africa: 1998-2001. Virus Res. 103:61-66. - PubMed
-
- Burland, T. G. 2000. DNASTAR's Lasergene sequence analysis software. Methods Mol. Biol. 132:71-91. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

