Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp
- PMID: 15322001
- PMCID: PMC517479
- DOI: 10.1128/IAI.72.9.5080-5088.2004
Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp
Abstract
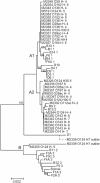
Enteroinvasive Escherichia coli (EIEC), a distinctive pathogenic form of E. coli causing dysentery, is similar in many properties to bacteria placed in the four species of Shigella. Shigella has been separated as a genus but in fact comprises several clones of E. coli. The evolutionary relationships of 32 EIEC strains of 12 serotypes have been determined by sequencing of four housekeeping genes and two plasmid genes which were used previously to determine the relationships of Shigella strains. The EIEC strains were grouped in four clusters with one outlier strain, indicating independent derivation of EIEC several times. Three of the four clusters contain more than one O antigen type. One EIEC strain (an O112ac:H- strain) was found in Shigella cluster 3 but is not identical to the Shigella cluster 3 D2 and B15 strains with the same O antigen. Two forms of the virulence plasmid pINV have been identified in Shigella strains by using the sequences of ipgD and mxiA genes, and all but two of our EIEC strains have pINV A. The EIEC strains were grouped in two subclusters with a very low level of variation, generally not intermingled with Shigella pINV A strains. The EIEC clusters based on housekeeping genes were reflected in the plasmid gene sequences, with some exceptions. Two strains were found in the pINV B form by using the ipgD sequence, with one strain having an mxiA sequence similar to the divergent sequence of D1. Clearly, EIEC and Shigella spp. form a pathovar of E. coli.
Figures



References
-
- Bando, S. Y., G. R. F. do Valle, M. B. Martinez, L. R. Trabulsi, and C. A. Moreira-Filho. 1998. Characterization of enteroinvasive Escherichia coli and Shigella strains by RAPD analysis. FEMS Microbiol. Lett. 165:159-165. - PubMed
-
- Bopp, C. A., F. W. Brenner, P. I. Fields, J. G. Wells, and N. A. Strockbine. 2003. Escherichia, Shigella, and Salmonella, p. 654-671. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, D.C.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

