Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus
- PMID: 19948792
- PMCID: PMC2798331
- DOI: 10.1105/tpc.109.068825
Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus
Abstract
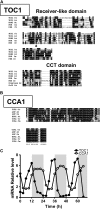
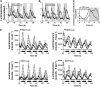
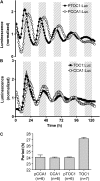
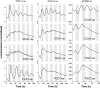
Biological rhythms that allow organisms to adapt to the solar cycle are generated by endogenous circadian clocks. In higher plants, many clock components have been identified and cellular rhythmicity is thought to be driven by a complex transcriptional feedback circuitry. In the small genome of the green unicellular alga Ostreococcus tauri, two of the master clock genes Timing of Cab expression1 (TOC1) and Circadian Clock-Associated1 (CCA1) appear to be conserved, but others like Gigantea or Early-Flowering4 are lacking. Stably transformed luciferase reporter lines and tools for gene functional analysis were therefore developed to characterize clock gene function in this simple eukaryotic system. This approach revealed several features that are comparable to those in higher plants, including the circadian regulation of TOC1, CCA1, and the output gene Chlorophyll a/b Binding under constant light, the relative phases of TOC1/CCA1 expression under light/dark cycles, arrhythmic overexpression phenotypes under constant light, the binding of CCA1 to a conserved evening element in the TOC1 promoter, as well as the requirement of the evening element for circadian regulation of TOC1 promoter activity. Functional analysis supports TOC1 playing a central role in the clock, but repression of CCA1 had no effect on clock function in constant light, arguing against a simple TOC1 /CCA1 one-loop clock in Ostreococcus. The emergence of functional genomics in a simple green cell with a small genome may facilitate increased understanding of how complex cellular processes such as the circadian clock have evolved in plants.
Figures


Comment in
-
Insights into the regulation of the core clock component TOC1 in the green picoeukaryote Ostreococcus.Plant Signal Behav. 2010 Mar;5(3):332-5. doi: 10.4161/psb.5.3.11212. Epub 2010 Mar 14. Plant Signal Behav. 2010. PMID: 20200488 Free PMC article.
References
-
- Alabadi, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Mas, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883. - PubMed
-
- Alabadi, D., Yanovsky, M.J., Mas, P., Harmer, S.L., and Kay, S.A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12 757–761. - PubMed
-
- Carre, I.A., and Kim, J.Y. (2002). MYB transcription factors in the Arabidopsis circadian clock. J. Exp. Bot. 53 1551–1557. - PubMed
-
- Courties, C., Vaquer, A., Troussellier, M., Lautier, J., Chrétiennot-Dinet, M.J., Neveux, J., Machado, C., and Claustre, H. (1994). Smallest eukaryotic organism. Nature 370 255.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

