Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotrophic gamma-proteobacteria in the Arabian sea
- PMID: 15812041
- PMCID: PMC1082540
- DOI: 10.1128/AEM.71.4.2079-2085.2005
Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotrophic gamma-proteobacteria in the Arabian sea
Abstract
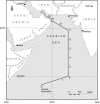
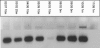
The spatial distribution of an uncultured clade of marine diazotrophic gamma-proteobacteria in the Arabian Sea was investigated by the development of a specific primer pair to amplify an internal fragment of nifH by PCR. These organisms were most readily detected in highly oligotrophic surface waters but could also be found in deeper waters below the nutricline. nifH transcripts originating from this clade were detected in oligotrophic surface waters and, in addition, in the deeper and the more productive near-coastal waters. The nifH sequences most closely related to the unidentified marine bacterial group are from environmental clones amplified from the Atlantic and Pacific Oceans. These findings suggest that these gamma-proteobacteria are widespread and likely to be an important component of the heterotrophic diazotrophic microbial community of the tropical and subtropical oceans.
Figures





References
-
- Capone, D. G., J. P. Zehr, H. W. Paerl, B. Bergman, and E. J. Carpenter. 1997. Trichodesmium, a globally significant marine cyanobacterium. Science 276:1221-1229.
-
- Carpenter, E. J., and K. Romans. 1991. Major role of the cyanobacterium Trichodesmium in nutrient cycling in the North Atlantic Ocean. Science 254:1356-1358. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

