Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea
- PMID: 15965264
- PMCID: PMC1456511
- DOI: 10.1534/genetics.104.033597
Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea
Abstract
Drosophila yakuba is widely distributed in sub-Saharan Africa, while D. santomea is endemic to the volcanic island of São Tomé in the Atlantic Ocean, 280 km west of Gabon. On São Tomé, D. yakuba is found mainly in open lowland forests, and D. santomea is restricted to the wet misty forests at higher elevations. At intermediate elevations, the species form a hybrid zone where hybrids occur at a frequency of approximately 1%. To determine the extent of gene flow between these species we studied polymorphism and divergence patterns in 29 regions distributed throughout the genome, including mtDNA and three genes on the Y chromosome. This multilocus approach, together with the comparison to the two allopatric species D. mauritiana and D. sechellia, allowed us to distinguish between forces that should affect all genes and forces that should act on some genes (e.g., introgression). Our results show that D. yakuba mtDNA has replaced that of D. santomea and that there is also significant introgression for two nuclear genes, yellow and salr. The majority of genes, however, has remained distinct. These two species therefore do not form a "hybrid swarm" in which much of the genome shows substantial introgression while disruptive selection maintains distinctness for only a few traits (e.g., pigmentation and male genitalia).
Figures
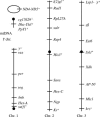
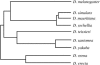
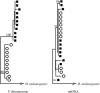
References
-
- Arnold, M. L., 1997. Natural Hybridization and Evolution. Oxford University Press, Oxford.
-
- Ashburner, M., 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Ballard, J. W., 2004. Sequential evolution of a symbiont inferred from the host: Wolbachia and Drosophila simulans. Mol. Biol. Evol. 21: 428–442. - PubMed
-
- Ballard, J. W. O., and M. C. Whitlock, 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13: 729–744. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

