HIV-1 recombinants with multiple parental strains in low-prevalence, remote regions of Cameroon: evolutionary relics?
- PMID: 20426823
- PMCID: PMC2879232
- DOI: 10.1186/1742-4690-7-39
HIV-1 recombinants with multiple parental strains in low-prevalence, remote regions of Cameroon: evolutionary relics?
Abstract
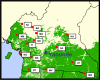
Background: The HIV pandemic disseminated globally from Central West Africa, beginning in the second half of the twentieth century. To elucidate the virologic origins of the pandemic, a cross-sectional study was conducted of the genetic diversity of HIV-1 strains in villagers in 14 remote locations in Cameroon and in hospitalized and STI patients. DNA extracted from PBMC was PCR amplified from HIV(+) subjects. Partial pol amplicons (N = 164) and nearly full virus genomes (N = 78) were sequenced. Among the 3956 rural villagers studied, the prevalence of HIV infection was 4.9%; among the hospitalized and clinic patients, it was 8.6%.
Results: Virus genotypes fell into two distinctive groups. A majority of the genotyped strains (109/164) were the circulating recombinant form (CRF) known to be endemic in West Africa and Central West Africa, CRF02_AG. The second most common genetic form (9/164) was the recently described CRF22_01A1, and the rest were a collection of 4 different subtypes (A2, D, F2, G) and 6 different CRFs (-01, -11, -13, -18, -25, -37). Remarkably, 10.4% of HIV-1 genomes detected (17/164) were heretofore undescribed unique recombinant forms (URF) present in only a single person. Nearly full genome sequencing was completed for 78 of the viruses of interest. HIV genetic diversity was commonplace in rural villages: 12 villages each had at least one newly detected URF, and 9 villages had two or more.
Conclusions: These results show that while CRF02_AG dominated the HIV strains in the rural villages, the remainder of the viruses had tremendous genetic diversity. Between the trans-species transmission of SIVcpz and the dispersal of pandemic HIV-1, there was a time when we hypothesize that nascent HIV-1 was spreading, but only to a limited extent, recombining with other local HIV-1, creating a large variety of recombinants. When one of those recombinants began to spread widely (i.e. became epidemic), it was recognized as a subtype. We hypothesize that the viruses in these remote Cameroon villages may represent that pre-epidemic stage of viral evolution.
Figures




References
-
- Van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, Shaw KS, Takehisa J, Kraus MH, Loul S, Butel C, Liegeois F, Yangda B, Sharp PM, Mpoudi-Ngole E, Delaporte E, Hahn BH, Peeters M. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368:155–171. doi: 10.1016/j.virol.2007.06.018. - DOI - PubMed
-
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. - DOI - PMC - PubMed
-
- Torimiro J, D'Arrigo R, Takou D, Nanfack A, Pizzi D, Ngong I, Carr JK, Joseph FP, Perno C-F, Cappelli G. Human Immunodeficiency Virus Type 1 Intersubtype Recombinants Predominate in the AIDS Epidemic in Cameroon. New Microbiologica. 2009;32:319–329. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

