Tests of parallel molecular evolution in a long-term experiment with Escherichia coli
- PMID: 16751270
- PMCID: PMC1482574
- DOI: 10.1073/pnas.0602917103
Tests of parallel molecular evolution in a long-term experiment with Escherichia coli
Abstract
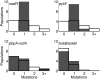
The repeatability of evolutionary change is difficult to quantify because only a single outcome can usually be observed for any precise set of circumstances. In this study, however, we have quantified the frequency of parallel and divergent genetic changes in 12 initially identical populations of Escherichia coli that evolved in identical environments for 20,000 cell generations. Unlike previous analyses in which candidate genes were identified based on parallel phenotypic changes, here we sequenced four loci (pykF, nadR, pbpA-rodA, and hokB/sokB) in which mutations of unknown effect had been discovered in one population, and then we compared the substitution pattern in these "blind" candidate genes with the pattern found in 36 randomly chosen genes. Two candidate genes, pykF and nadR, had substitutions in all 11 other populations, and the other 2 in several populations. There were very few cases, however, in which the exact same mutations were substituted, in contrast to the findings from conceptually related work performed with evolving virus populations. No random genes had any substitutions except in four populations that evolved defects in DNA repair. Tests of four different statistical aspects of the pattern of molecular evolution all indicate that adaptation by natural selection drove the parallel changes in these candidate genes.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
-
- Simpson G. G. The Major Features of Evolution. New York: Columbia Univ. Press; 1953.
-
- Harvey P. H., Pagel M. The Comparative Method in Evolutionary Biology. Oxford: Oxford Univ. Press; 1991.
-
- Futuyma D. J. Evolutionary Biology. Sunderland, MA: Sinauer; 1998.
-
- Losos J. B., Jackman T. R., Larson A., de Queiroz K., Rodríguez-Schettino L. Science. 1998;279:2115–2118. - PubMed
-
- Rundle H. D., Nagel L., Boughman J. W., Schluter D. Science. 2000;287:306–308. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

