The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk
- PMID: 15826937
- PMCID: PMC1464088
- DOI: 10.1074/jbc.M501636200
The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk
Abstract
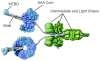
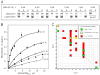
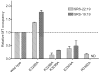
The microtubule-binding domain (MTBD) of dynein is separated from the AAA (ATPase with any other activity) core of the motor by an approximately 15-nm stalk that is predicted to consist of an antiparallel coiled coil. However, the structure of this coiled coil and the mechanism it uses to mediate communication between the MTBD and ATP-binding core are unknown. Here, we sought to identify the optimal alignment between the hydrophobic heptad repeats in the two strands of the stalk coiled coil. To do this, we fused the MTBD of mouse cytoplasmic dynein, together with 12-36 residues of its stalk, onto a stable coiled-coil base provided by Thermus thermophilus seryl-tRNA synthetase and tested these chimeric constructs for microtubule binding in vitro. The results identified one alignment that yielded a protein displaying high affinity for microtubules (2.2 microM). The effects of mutations applied to the MTBD of this construct paralleled those previously reported (Koonce, M. P., and Tikhonenko, I. (2000) Mol. Biol. Cell 11, 523-529) for an intact dynein motor unit in the absence of ATP, suggesting that it resembles the tight binding state of native intact dynein. All other alignments showed at least 10-fold lower affinity for microtubules with the exception of one, which had an intermediate affinity. Based on these results and on amino acid sequence analysis, we hypothesize that dynein utilizes small amounts of sliding displacement between the two strands of its coiled-coil stalk as a means of communication between the AAA core of the motor and the MTBD during the mechanochemical cycle.
Figures

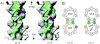
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

