Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome
- PMID: 16157677
- PMCID: PMC1456821
- DOI: 10.1534/genetics.105.043976
Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome
Abstract
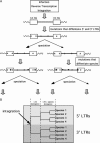
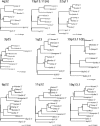
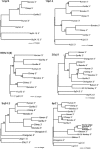
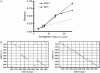
HERV elements make up a significant fraction of the human genome and, as interspersed repetitive elements, have the capacity to provide substrates for ectopic recombination and gene conversion events. To understand the extent to which these events occur and gain further insight into the complex evolutionary history of these elements in our genome, we undertook a phylogenetic study of the long terminal repeat sequences of 15 HERV-K(HML-2) elements in various primate species. This family of human endogenous retroviruses first entered the primate genome between 35 and 45 million years ago. Throughout primate evolution, these elements have undergone bursts of amplification. From this analysis, which is the largest-scale study of HERV sequence dynamics during primate evolution to date, we were able to detect intraelement gene conversion and recombination at five HERV-K loci. We also found evidence for replacement of an ancient element by another HERV-K provirus, apparently reflecting an occurrence of retroviral integration by homologous recombination. The high frequency of these events casts doubt on the accuracy of integration time estimates based only on divergence between retroelement LTRs.
Figures



References
-
- Balding, D. J., R. A. Nichols and D. M. Hunt, 1992. Detecting gene conversion: primate visual pigment genes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 249: 275–280. - PubMed
-
- Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd et al., 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9: 861–868. - PubMed
-
- Belshaw, R., A. Katzourakis, J. Paces, A. Burt and M. Tristem, 2005. High copy number in human endogenous retrovirus (HERV) families is associated with copying mechanisms in addition to re-infection. Mol. Biol. Evol. 22: 814–817. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

