Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays
- PMID: 15998911
- PMCID: PMC1172043
- DOI: 10.1101/gr.3455305
Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays
Abstract
Recently, we mapped the sites of transcription across approximately 30% of the human genome and elucidated the structures of several hundred novel transcripts. In this report, we describe a novel combination of techniques including the rapid amplification of cDNA ends (RACE) and tiling array technologies that was used to further characterize transcripts in the human transcriptome. This technical approach allows for several important pieces of information to be gathered about each array-detected transcribed region, including strand of origin, start and termination positions, and the exonic structures of spliced and unspliced coding and noncoding RNAs. In this report, the structures of transcripts from 14 transcribed loci, representing both known genes and unannotated transcripts taken from the several hundred randomly selected unannotated transcripts described in our previous work are represented as examples of the complex organization of the human transcriptome. As a consequence of this complexity, it is not unusual that a single base pair can be part of an intricate network of multiple isoforms of overlapping sense and antisense transcripts, the majority of which are unannotated. Some of these transcripts follow the canonical splicing rules, whereas others combine the exons of different genes or represent other types of noncanonical transcripts. These results have important implications concerning the correlation of genotypes to phenotypes, the regulation of complex interlaced transcriptional patterns, and the definition of a gene.
Figures

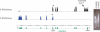



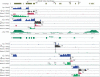

References
-
- Bijlsma, M.F., Spek, C.A., and Peppelenbosch, M.P. 2004. Hedgehog: An unusual signal transducer. Bioessays 26: 387-394. - PubMed
-
- Cawley, S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499-509. - PubMed
-
- Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science http://www.sciencemag.org/cgi/content/abstract/1108625v2. - PubMed
-
- Hawkins, P.R., Jin, P., and Fu, G.K. 2003. Full-length cDNA synthesis for long-distance RT-PCR of large mRNA transcripts. Biotechniques 34: 768-770, 772-773. - PubMed
Web site references
-
- http://transcriptome.affymetrix.com/publication/race/; RACE/array data of this paper.
-
- http://www.affymetrix.com/support/developer/downloads/TilingArrayTools/i...; Integrated Genome Browser.
-
- http://www.genome.ucsc.edu; UCSC Genome Browser.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
