VSG switching in Trypanosoma brucei: antigenic variation analysed using RNAi in the absence of immune selection
- PMID: 16135228
- PMCID: PMC1618954
- DOI: 10.1111/j.1365-2958.2005.04795.x
VSG switching in Trypanosoma brucei: antigenic variation analysed using RNAi in the absence of immune selection
Abstract
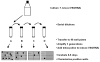
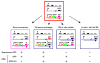
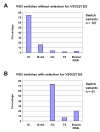
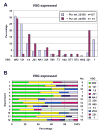
Trypanosoma brucei relies on antigenic variation of its variant surface glycoprotein (VSG) coat for survival. We show that VSG switching can be efficiently studied in vitro using VSG RNAi in place of an immune system to select for switch variants. Contrary to models predicting an instant switch after inhibition of VSG synthesis, switching was not induced by VSG RNAi and occurred at a rate of 10(-4) per division. We find a highly reproducible hierarchy of VSG activation, which appears to be capable of resetting, whereby more than half of the switch events over 12 experiments were to one of two VSGs. We characterized switched clones according to switch mechanism using marker genes in the active VSG expression site (ES). Transcriptional switches between ESs were the preferred switching mechanism, whereby at least 10 of the 17 ESs identified in T. brucei 427 can be functionally active in vitro. We could specifically select for switches mediated by DNA rearrangements by inducing VSG RNAi in the presence of drug selection for the active ES. Most of the preferentially activated VSGs could be activated by multiple mechanisms. This VSG RNAi-based procedure provides a rapid and powerful means for analysing VSG switching in African trypanosomes entirely in vitro.
Figures



References
-
- Aline RF, Myler PJ, Stuart KD. Trypanosoma brucei: frequent loss of a telomeric variant surface glycoprotein gene. Exp Parasitol. 1989;68:8–16. - PubMed
-
- Bangs JD, Uyetake L, Brickman MJ, Balber AE, Boothroyd JC. Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J Cell Sci. 1993;105:1101–1113. - PubMed
-
- Barry JD. Antigenic variation during Trypanosoma vivax infections of different host species. Parasitology. 1986;92:51–65. - PubMed
-
- Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol. 2001;49:1–70. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

