Neutral evolution of the nonbinding region of the anthocyanin regulatory gene Ipmyb1 in Ipomoea
- PMID: 15944366
- PMCID: PMC1449781
- DOI: 10.1534/genetics.104.034975
Neutral evolution of the nonbinding region of the anthocyanin regulatory gene Ipmyb1 in Ipomoea
Abstract
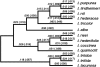
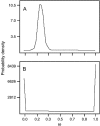
Plant transcription factors often contain domains that evolve very rapidly. Although it has been suggested that this rapid evolution may contribute substantially to phenotypic differentiation among species, this suggestion has seldom been tested explicitly. We tested the validity of this hypothesis by examining the rapidly evolving non-DNA-binding region of an R2R3-myb transcription factor that regulates anthocyanin expression in flowers of the genus Ipomoea. We first provide evidence that the W locus in Ipomoea purpurea, which determines whether flowers will be pigmented or white, corresponds to a myb gene segregating in southeastern U.S. populations for one functional allele and one nonfunctional allele. While the binding domain exhibits substantial selective constraint, the nonbinding region evolves at an average K(a)/K(s) ratio of 0.74. This elevated rate of evolution is due to relaxed constraint rather than to increased levels of positive selection. Despite this relaxed constraint, however, approximately 20-25% of the codons, randomly distributed throughout the nonbinding region, are highly constrained, with the remainder evolving neutrally, indicating that the entire region performs important function(s). Our results provide little indication that rapid evolution in this regulatory gene is driven by natural selection or that it is responsible for floral-color differences among Ipomoea species.
Figures




References
-
- Arthur, W., 2002. The emerging conceptual framework of evolutionary developmental biology. Nature 415: 757–764. - PubMed
-
- Cherry, L. M., S. M. Case and A. C. Wilson, 1978. Frog perspective on the morphological difference between humans and chimpanzees. Science 200: 209–211. - PubMed
-
- Cubas, P., C. Vincent and E. Coen, 1999. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

