Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery
- PMID: 27428751
- PMCID: PMC5019950
- DOI: 10.1038/ng.3592
Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery
Abstract
The Greater Middle East (GME) has been a central hub of human migration and population admixture. The tradition of consanguinity, variably practiced in the Persian Gulf region, North Africa, and Central Asia, has resulted in an elevated burden of recessive disease. Here we generated a whole-exome GME variome from 1,111 unrelated subjects. We detected substantial diversity and admixture in continental and subregional populations, corresponding to several ancient founder populations with little evidence of bottlenecks. Measured consanguinity rates were an order of magnitude above those in other sampled populations, and the GME population exhibited an increased burden of runs of homozygosity (ROHs) but showed no evidence for reduced burden of deleterious variation due to classically theorized 'genetic purging'. Applying this database to unsolved recessive conditions in the GME population reduced the number of potential disease-causing variants by four- to sevenfold. These results show variegated genetic architecture in GME populations and support future human genetic discoveries in Mendelian and population genetics.
Conflict of interest statement
The authors declare no competing financial interests
Figures
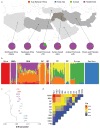
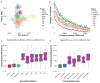
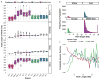
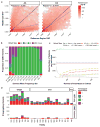
Comment in
-
Genomic landscape of the Greater Middle East.Nat Genet. 2016 Aug 30;48(9):978-9. doi: 10.1038/ng.3652. Nat Genet. 2016. PMID: 27573686
References
-
- Anwar WA, Khyatti M, Hemminki K. Consanguinity and genetic diseases in North Africa and immigrants to Europe. Eur J Public Health. 2014;24(Suppl 1):57–63. - PubMed
-
- Hussain R, Bittles AH. The prevalence and demographic characteristics of consanguineous marriages in Pakistan. J Biosoc Sci. 1998;30:261–75. - PubMed
-
- Sheffield VC, Stone EM, Carmi R. Use of isolated inbred human populations for identification of disease genes. Trends Genet. 1998;14:391–6. - PubMed
-
- Sharp JM. The Broader Middle East and North Africa Initiative: An overview. CRS Report for Congress; 2005.
MeSH terms
Substances
Grants and funding
- UL1 TR000043/TR/NCATS NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U01 AI088685/AI/NIAID NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- UL1 TR001866/TR/NCATS NIH HHS/United States
- R01 NS048453/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 AI095983/AI/NIAID NIH HHS/United States
- U01 AI109697/AI/NIAID NIH HHS/United States
- R21 AI107508/AI/NIAID NIH HHS/United States
- U54 HG006504/HG/NHGRI NIH HHS/United States
- UL1 RR024143/RR/NCRR NIH HHS/United States
- P01 HD070494/HD/NICHD NIH HHS/United States
- R01 AI088364/AI/NIAID NIH HHS/United States
- P01 AI061093/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

