Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes
- PMID: 17989253
- PMCID: PMC2099591
- DOI: 10.1101/gr.6679507
Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes
Abstract
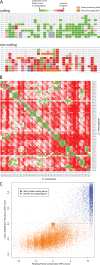
The availability of sequenced genomes from 12 Drosophila species has enabled the use of comparative genomics for the systematic discovery of functional elements conserved within this genus. We have developed quantitative metrics for the evolutionary signatures specific to protein-coding regions and applied them genome-wide, resulting in 1193 candidate new protein-coding exons in the D. melanogaster genome. We have reviewed these predictions by manual curation and validated a subset by directed cDNA screening and sequencing, revealing both new genes and new alternative splice forms of known genes. We also used these evolutionary signatures to evaluate existing gene annotations, resulting in the validation of 87% of genes lacking descriptive names and identifying 414 poorly conserved genes that are likely to be spurious predictions, noncoding, or species-specific genes. Furthermore, our methods suggest a variety of refinements to hundreds of existing gene models, such as modifications to translation start codons and exon splice boundaries. Finally, we performed directed genome-wide searches for unusual protein-coding structures, discovering 149 possible examples of stop codon readthrough, 125 new candidate ORFs of polycistronic mRNAs, and several candidate translational frameshifts. These results affect >10% of annotated fly genes and demonstrate the power of comparative genomics to enhance our understanding of genome organization, even in a model organism as intensively studied as Drosophila melanogaster.
Figures




References
-
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Andrews J., Smith M., Merakovsky J., Coulson M., Hannan F., Kelly L.E., Smith M., Merakovsky J., Coulson M., Hannan F., Kelly L.E., Merakovsky J., Coulson M., Hannan F., Kelly L.E., Coulson M., Hannan F., Kelly L.E., Hannan F., Kelly L.E., Kelly L.E. The stoned locus of Drosophila melanogaster produces a dicistronic transcript and encodes two distinct polypeptides. Genetics. 1996;143:1699–1711. - PMC - PubMed
-
- Bergman C.M., Pfeiffer B.D., Rincon-Limas D.E., Hoskins R.A., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Pfeiffer B.D., Rincon-Limas D.E., Hoskins R.A., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Rincon-Limas D.E., Hoskins R.A., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Hoskins R.A., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Wang A.M., Kronmiller B., Pacleb J., Park S., Kronmiller B., Pacleb J., Park S., Pacleb J., Park S., Park S., et al. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0086. - DOI - PMC - PubMed
-
- Bergstrom D.E., Merli C.A., Cygan J.A., Shelby R., Blackman R.K., Merli C.A., Cygan J.A., Shelby R., Blackman R.K., Cygan J.A., Shelby R., Blackman R.K., Shelby R., Blackman R.K., Blackman R.K. Regulatory autonomy and molecular characterization of the Drosophila out at first gene. Genetics. 1995;139:1331–1346. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
