dsMTL: a computational framework for privacy-preserving, distributed multi-task machine learning
- PMID: 36073911
- PMCID: PMC9620828
- DOI: 10.1093/bioinformatics/btac616
dsMTL: a computational framework for privacy-preserving, distributed multi-task machine learning
Abstract
Motivation: In multi-cohort machine learning studies, it is critical to differentiate between effects that are reproducible across cohorts and those that are cohort-specific. Multi-task learning (MTL) is a machine learning approach that facilitates this differentiation through the simultaneous learning of prediction tasks across cohorts. Since multi-cohort data can often not be combined into a single storage solution, there would be the substantial utility of an MTL application for geographically distributed data sources.
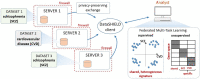
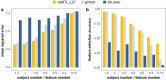
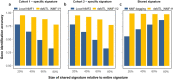
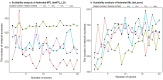
Results: Here, we describe the development of 'dsMTL', a computational framework for privacy-preserving, distributed multi-task machine learning that includes three supervised and one unsupervised algorithms. First, we derive the theoretical properties of these methods and the relevant machine learning workflows to ensure the validity of the software implementation. Second, we implement dsMTL as a library for the R programming language, building on the DataSHIELD platform that supports the federated analysis of sensitive individual-level data. Third, we demonstrate the applicability of dsMTL for comorbidity modeling in distributed data. We show that comorbidity modeling using dsMTL outperformed conventional, federated machine learning, as well as the aggregation of multiple models built on the distributed datasets individually. The application of dsMTL was computationally efficient and highly scalable when applied to moderate-size (n < 500), real expression data given the actual network latency.
Availability and implementation: dsMTL is freely available at https://github.com/transbioZI/dsMTLBase (server-side package) and https://github.com/transbioZI/dsMTLClient (client-side package).
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures
References
-
- Cao H. et al. (2019) RMTL: an R library for multi-task learning. Bioinformatics, 35, 1797–1798. - PubMed
-
- Consotia T.D. (2019). Community packages of DataSHIELD. https://www.datashield.org/help/community-packages.

