Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04
- PMID: 19376856
- PMCID: PMC2698493
- DOI: 10.1128/JB.00155-09
Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04
Abstract
Bifidobacteria are important members of the human gut flora, especially in infants. Comparative genomic analysis of two Bifidobacterium animalis subsp. lactis strains revealed evolution by internal deletion of consecutive spacer-repeat units within a novel clustered regularly interspaced short palindromic repeat locus, which represented the largest differential content between the two genomes. Additionally, 47 single nucleotide polymorphisms were identified, consisting primarily of nonsynonymous mutations, indicating positive selection and/or recent divergence. A particular nonsynonymous mutation in a putative glucose transporter was linked to a negative phenotypic effect on the ability of the variant to catabolize glucose, consistent with a modification in the predicted protein transmembrane topology. Comparative genome sequence analysis of three Bifidobacterium species provided a core genome set of 1,117 orthologs complemented by a pan-genome of 2,445 genes. The genome sequences of the intestinal bacterium B. animalis subsp. lactis provide insights into rapid genome evolution and the genetic basis for adaptation to the human gut environment, notably with regard to catabolism of dietary carbohydrates, resistance to bile and acid, and interaction with the intestinal epithelium. The high degree of genome conservation observed between the two strains in terms of size, organization, and sequence is indicative of a genomically monomorphic subspecies and explains the inability to differentiate the strains by standard techniques such as pulsed-field gel electrophoresis.
Figures
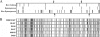

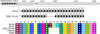
References
-
- Ajdić, D., W. M. McShan, R. E. McLaughlin, G. Savić, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. - PMC - PubMed
-
- Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. D. W. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 1023906-3912. - PMC - PubMed
-
- Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. - PubMed
-
- Bartosch, S., E. J. Woodmansey, J. C. M. Paterson, M. E. T. McMurdo, and G. T. Macfarlane. 2005. Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real-time polymerase chain reaction and counting of viable bacteria. Clin. Infect. Dis. 4028-37. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

