Genomes of Ashbya fungi isolated from insects reveal four mating-type loci, numerous translocations, lack of transposons, and distinct gene duplications
- PMID: 23749448
- PMCID: PMC3737163
- DOI: 10.1534/g3.112.002881
Genomes of Ashbya fungi isolated from insects reveal four mating-type loci, numerous translocations, lack of transposons, and distinct gene duplications
Abstract
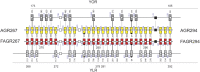
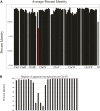
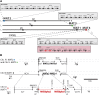
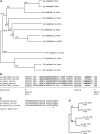
The filamentous fungus Ashbya gossypii is a cotton pathogen transmitted by insects. It is readily grown and manipulated in the laboratory and is commercially exploited as a natural overproducer of vitamin B2. Our previous genome analysis of A. gossypii isolate ATCC10895, collected in Trinidad nearly 100 years ago, revealed extensive synteny with the Saccharomyces cerevisiae genome, leading us to use it as a model organism to understand the evolution of filamentous growth. To further develop Ashbya as a model system, we have investigated the ecological niche of A. gossypii and isolated additional strains and a sibling species, both useful in comparative analysis. We isolated fungi morphologically similar to A. gossypii from different plant-feeding insects of the suborder Heteroptera, generated a phylogenetic tree based on rDNA-ITS sequences, and performed high coverage short read sequencing with one A. gossypii isolate from Florida, a new species, Ashbya aceri, isolated in North Carolina, and a genetically marked derivative of ATCC10895 intensively used for functional studies. In contrast to S. cerevisiae, all strains carry four not three mating type loci, adding a new puzzle in the evolution of Ashbya species. Another surprise was the genome identity of 99.9% between the Florida strain and ATCC10895, isolated in Trinidad. The A. aceri and A. gossypii genomes show conserved gene orders rearranged by eight translocations, 90% overall sequence identity, and fewer tandem duplications in the A. aceri genome. Both species lack transposable elements. Finally, our work identifies plant-feeding insects of the suborder Heteroptera as the most likely natural reservoir of Ashbya, and that infection of cotton and other plants may be incidental to the growth of the fungus in its insect host.
Keywords: fungal ecology; intron evolution; mating type; tandem duplications.
Figures


References
-
- Altmann-Jöhl R., Philippsen P., 1996. AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol. Gen. Genet. 250: 69–80 - PubMed
-
- Ashby S. F., 1916. Annual Report of the Microbiologist, 1915–1916. Annual report of the Department of Agriculture, Jamaica, pp. 29–31
-
- Ashby S. F., Nowell W., 1926. The fungi of Stigmatomycosis. Ann. Bot. (Lond.) 40: 69–83
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
