Enterobacter oligotrophica sp. nov., a novel oligotroph isolated from leaf soil
- PMID: 31066221
- PMCID: PMC7650607
- DOI: 10.1002/mbo3.843
Enterobacter oligotrophica sp. nov., a novel oligotroph isolated from leaf soil
Abstract
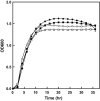
A novel oligotrophic bacterium, designated strain CCA6, was isolated from leaf soil collected in Japan. Cells of the strain were found to be a Gram-negative, non-sporulating, motile, rod-shaped bacterium. Strain CCA6 grew at 10-45°C (optimum 20°C) and pH 4.5-10.0 (optimum pH 5.0). The strain was capable of growth in poor-nutrient (oligotrophic) medium, and growth was unaffected by high-nutrient medium. The major fatty acid and predominant quinone system were C16:0 and ubiquinone-8. Phylogenetic analysis based on 16S rRNA gene sequences indicated strain CCA6 presented as a member of the family Enterobacteriaceae. Multilocus sequence analysis (MLSA) based on fragments of the atpD, gyrB, infB, and rpoB gene sequences was performed to further identify strain CCA6. The MLSA showed clear branching of strain CCA6 with respect to Enterobacter type strains. The complete genome of strain CCA6 consisted of 4,476,585 bp with a G+C content of 54.3% and comprising 4,372 predicted coding sequences. The genome average nucleotide identity values between strain CCA6 and the closest related Enterobacter type strain were <88.02%. Based on its phenotypic, chemotaxonomic and phylogenetic features, strain CCA6 (=HUT 8142T =KCTC 62525T ) can be considered as a novel species within the genus Enterobacter with the proposed name Enterobacter oligotrophica.
Keywords: Enterobacter; Voges-Proskauer test; average nucleotide identity value analysis; genome sequence; multilocus sequence analysis; oligotroph.
© 2019 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Brady, C. , Cleenwerck, I. , Venter, S. , Coutinho, T. , & De Vos, P. (2013). Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter . Systematic and Applied Microbiology., 36, 309–319. 10.1016/j.syapm.2013.03.005 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

