Genomics and host specialization of honey bee and bumble bee gut symbionts
- PMID: 25053814
- PMCID: PMC4128107
- DOI: 10.1073/pnas.1405838111
Genomics and host specialization of honey bee and bumble bee gut symbionts
Abstract
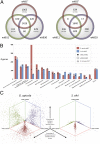
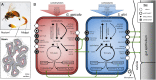
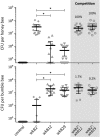
Gilliamella apicola and Snodgrassella alvi are dominant members of the honey bee (Apis spp.) and bumble bee (Bombus spp.) gut microbiota. We generated complete genomes of the type strains G. apicola wkB1(T) and S. alvi wkB2(T) (isolated from Apis), as well as draft genomes for four other strains from Bombus. G. apicola and S. alvi were found to occupy very different metabolic niches: The former is a saccharolytic fermenter, whereas the latter is an oxidizer of carboxylic acids. Together, they may form a syntrophic network for partitioning of metabolic resources. Both species possessed numerous genes [type 6 secretion systems, repeats in toxin (RTX) toxins, RHS proteins, adhesins, and type IV pili] that likely mediate cell-cell interactions and gut colonization. Variation in these genes could account for the host fidelity of strains observed in previous phylogenetic studies. Here, we also show the first experimental evidence, to our knowledge, for this specificity in vivo: Strains of S. alvi were able to colonize their native bee host but not bees of another genus. Consistent with specific, long-term host association, comparative genomic analysis revealed a deep divergence and little or no gene flow between Apis and Bombus gut symbionts. However, within a host type (Apis or Bombus), we detected signs of horizontal gene transfer between G. apicola and S. alvi, demonstrating the importance of the broader gut community in shaping the evolution of any one member. Our results show that host specificity is likely driven by multiple factors, including direct host-microbe interactions, microbe-microbe interactions, and social transmission.
Keywords: bacterial genomics; strain variation; symbiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Oh PL, et al. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010;4(3):377–387. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

