Complete genome of a European hepatitis C virus subtype 1g isolate: phylogenetic and genetic analyses
- PMID: 18533988
- PMCID: PMC2438343
- DOI: 10.1186/1743-422X-5-72
Complete genome of a European hepatitis C virus subtype 1g isolate: phylogenetic and genetic analyses
Abstract
Background: Hepatitis C virus isolates have been classified into six main genotypes and a variable number of subtypes within each genotype, mainly based on phylogenetic analysis. Analyses of the genetic relationship among genotypes and subtypes are more reliable when complete genome sequences (or at least the full coding region) are used; however, so far 31 of 80 confirmed or proposed subtypes have at least one complete genome available. Of these, 20 correspond to confirmed subtypes of epidemic interest.
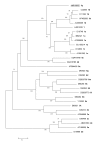
Results: We present and analyse the first complete genome sequence of a HCV subtype 1g isolate. Phylogenetic and genetic distance analyses reveal that HCV-1g is the most divergent subtype among the HCV-1 confirmed subtypes. Potential genomic recombination events between genotypes or subtype 1 genomes were ruled out. We demonstrate phylogenetic congruence of previously deposited partial sequences of HCV-1g with respect to our sequence.
Conclusion: In light of this, we propose changing the current status of its subtype-specific designation from provisional to confirmed.
Figures
References
-
- WHO Hepatitis C – global prevalence (update) Weekly Epidemiological Record. 1999;49:425–427. - PubMed
-
- Robertson B, Myers G, Howard C, Brettin T, Bukh J, Gaschen B, Gojobori T, Maertens G, Mizokami M, Nainan O, Netesov S, Nishioka K, Shin-I T, Simmonds P, Smith D, Stuyver L, Weiner A. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch Virol. 1998;143:2493–2503. doi: 10.1007/s007050050479. - DOI - PubMed
-
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin-I T, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. - DOI - PubMed
-
- Carrat F, Bani-Sadr F, Pol S, Rosenthal E, Lunel-Fabiani F, Benzekri A, Morand P, Goujard C, Pialoux G, Piroth L, Salmon-Céron D, Degott C, Cacoub P, Perronne C, ANRS HCO2 RIBAVIC Study Team Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. JAMA. 2004;292:2839–2848. doi: 10.1001/jama.292.23.2839. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

