Three putative cation/proton antiporters from the soda lake alkaliphile Alkalimonas amylolytica N10 complement an alkali-sensitive Escherichia coli mutant
- PMID: 17600061
- PMCID: PMC2538799
- DOI: 10.1099/mic.0.2007/007450-0
Three putative cation/proton antiporters from the soda lake alkaliphile Alkalimonas amylolytica N10 complement an alkali-sensitive Escherichia coli mutant
Abstract
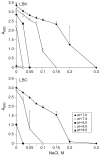
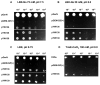
Attempts to identify members of the antiporter complement of the alkali- and saline-adapted soda lake alkaliphile Alkalimonas amylolytica N10 have used screens of DNA libraries in antiporter-deficient Escherichia coli KNabc. Earlier screens used Na(+) or Li(+) for selection but only identified one NhaD-type antiporter whose properties were inconsistent with a robust role in pH homeostasis. Here, new screens using elevated pH for selection identified three other putative antiporter genes that conferred resistance to pH >or=8.5 as well as Na(+) resistance. The three predicted gene products were in the calcium/cation antiporter (CaCA), cation/proton antiporter-2 (CPA2) and cation/proton antiporter-1 (CPA1) families of membrane transporters, and were designated Aa-CaxA, Aa-KefB and Aa-NhaP respectively, reflecting homology within those families. Aa-CaxA conferred the poorest Na(+) resistance and also conferred modest Ca(2+) resistance. Aa-KefB and Aa-NhaP inhibited growth of a K(+) uptake-deficient E. coli mutant (TK2420), suggesting that they catalysed K(+) efflux. For Aa-NhaP, the reversibility of the growth inhibition by high K(+) concentrations depended upon an organic nitrogen source, e.g. glutamine, rather than ammonium. This suggests that as well as K(+) efflux is catalysed by Aa-NhaP. Vesicles of E. coli KNabc expressing Aa-NhaP, which conferred the strongest alkali resistance, exhibited K(+)/H(+) antiport activity in a pH range from 7.5 to 9.5, and with an apparent K(m) for K(+) of 0.5 mM at pH 8.0. The properties of this antiporter are consistent with the possibility that this soda lake alkaliphile uses K(+)( )/H(+) antiport as part of its alkaline pH homeostasis mechanism and part of its capacity to reduce potentially toxic accumulation of cytoplasmic K(+) or respectively, under conditions of high osmolarity or active amino acid catabolism.
Figures

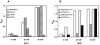

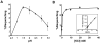
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Booth IR, Edwards MD, Murray E, Miller S. The role of bacterial channels in cell physiology. In: Kubalski A, Marinac B, editors. Bacterial Ion Channels and Their Eukaryotic Homologs. Washington, D.C.: ASM Press; 2005. pp. 291–312.
-
- Brockman RW, Heppel LA. On the localization of alkaline phosphatase and cyclic phosphodiesterase in Escherichia coli. Biochemistry. 1968;7:2554–2562. - PubMed
-
- Buurman ET, Teixeira de Mattos MJ, Neijssel OM. Futile cycling of ammonium ions via the high affinity potassium uptake system (Kdp) of Escherichia coli. Arch Microbiol. 1991;155:391–395. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

