A hypervariable N-terminal region of Yersinia LcrV determines Toll-like receptor 2-mediated IL-10 induction and mouse virulence
- PMID: 16239347
- PMCID: PMC1276055
- DOI: 10.1073/pnas.0504728102
A hypervariable N-terminal region of Yersinia LcrV determines Toll-like receptor 2-mediated IL-10 induction and mouse virulence
Abstract
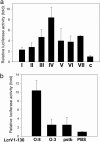
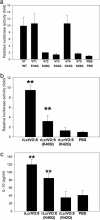
The virulence antigen LcrV of Yersinia enterocolitica O:8 induces IL-10 in macrophages via Toll-like receptor 2 (TLR2). The TLR2-active region of LcrV is localized within its N-terminal amino acids (aa) 31-57. Sequencing of codons 25-92 of the lcrV gene from 59 strains of the three pathogenic Yersinia species revealed a hypervariable hotspot within aa 40-61. According to these sequence differences, seven LcrV groups were identified, with Y. pestis and Y. pseudotuberculosis represented in group I and the other six distributed within Y. enterocolitica. By testing LcrV sequence-derived synthetic oligopeptides of all seven LcrV groups in CD14/TLR2-transfected human embryonic kidney 293 cells, we found the highest TLR2 activity with a peptide derived from group IV comprising exclusively Y. enterocolitica O:8 strains. These findings were verified in murine peritoneal macrophages by using recombinant LcrV truncates representing aa 1-130 from different Yersinia spp. By systematically replacing charged aa residues by glutamine in synthetic oligopeptides, we show that the K42Q substitution leads to abrogation of TLR2 activity in both in vitro cell systems. This K42Q substitution was introduced in the lcrV gene from Y. enterocolitica O:8 WA-C(pYV), resulting in WA-C(pYVLcrV(K42Q)), which turned out to be less virulent for C57BL/6 mice than the parental strain. This difference in virulence was not observed in TLR2(-/-) or IL-10(-/-) mice, proving that LcrV contributes to virulence by TLR2-mediated IL-10 induction. LcrV is a defined bacterial virulence factor shown to target the TLR system for evasion of the host's immune response.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

