Molecular evolution in the Drosophila melanogaster species subgroup: frequent parameter fluctuations on the timescale of molecular divergence
- PMID: 16387879
- PMCID: PMC1456288
- DOI: 10.1534/genetics.105.049676
Molecular evolution in the Drosophila melanogaster species subgroup: frequent parameter fluctuations on the timescale of molecular divergence
Abstract
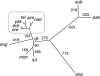
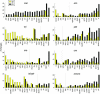
Although mutation, genetic drift, and natural selection are well established as determinants of genome evolution, the importance (frequency and magnitude) of parameter fluctuations in molecular evolution is less understood. DNA sequence comparisons among closely related species allow specific substitutions to be assigned to lineages on a phylogenetic tree. In this study, we compare patterns of codon usage and protein evolution in 22 genes (>11,000 codons) among Drosophila melanogaster and five relatives within the D. melanogaster subgroup. We assign changes to eight lineages using a maximum-likelihood approach to infer ancestral states. Uncertainty in ancestral reconstructions is taken into account, at least to some extent, by weighting reconstructions by their posterior probabilities. Four of the eight lineages show potentially genomewide departures from equilibrium synonymous codon usage; three are decreasing and one is increasing in major codon usage. Several of these departures are consistent with lineage-specific changes in selection intensity (selection coefficients scaled to effective population size) at silent sites. Intron base composition and rates and patterns of protein evolution are also heterogeneous among these lineages. The magnitude of forces governing silent, intron, and protein evolution appears to have varied frequently, and in a lineage-specific manner, within the D. melanogaster subgroup.
Figures
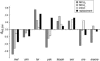

References
-
- Akashi, H., 2001. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11: 660–666. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

