Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species
- PMID: 16517861
- PMCID: PMC1393131
- DOI: 10.1128/JCM.44.3.827-832.2006
Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species
Abstract
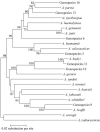
Acinetobacter species are defined on the basis of several phenotypic characters, results of DNA-DNA homology, and more recently, similarities or dissimilarities in 16S rRNA gene sequences. However, the 16S rRNA gene is not polymorphic enough to clearly distinguish all Acinetobacter species. We used an RNA polymerase beta-subunit gene (rpoB)-based identification scheme for the delineation of species within the genus Acinetobacter, and towards that end, we determined the complete rpoB gene and flanking spacer (rplL-rpoB and rpoB-rpoC) sequences of the 17 reference strains of Acinetobacter species and 7 unnamed genomospecies. By using complete gene sequences (4,089 bp), we clearly separated all species and grouped them into different clusters. A phylogenetic tree constructed using these sequences was supported by bootstrap values higher than those obtained with 16S rRNA or the gyrB or recA gene. Four pairs of primers enabled us to amplify and sequence two highly polymorphic partial sequences (350 and 450 bp) of the rpoB gene. These and flanking spacers were designed and tested for rapid identification of the 17 reference strains of Acinetobacter species and 7 unnamed genomospecies. Each of these four variable sequences enabled us to delineate most species. Sequences of at least two polymorphic sequences should be used to distinguish Acinetobacter grimontii, Acinetobacter junii, Acinetobacter baylyi, and genomic species 9 from one another. Finally, 21 clinical isolates of Acinetobacter baumannii were tested for intraspecies relationships and assigned correctly to the same species by comparing the partial sequences of the rpoB gene and its flanking spacers.
Figures


References
-
- Alexander, M., F. Ismail, P. J. H. Jackman, and W. C. Nobel. 1984. Fingerprinting Acinetobacter strains from clinical sources by numerical analysis of electrophoretic patterns. J. Med. Microbiol. 18:55-68. - PubMed
-
- Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. - PMC - PubMed
-
- Bouvet, P. J. M., and P. A. D. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36:228-240.
-
- Bouvet, P. J. M., and P. A. D. Grimont. 1987. Identification and biotyping of clinical isolates of Acinetobacter. Ann. Inst. Pasteur Microbiol. 138:569-578. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

