Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003
- PMID: 16272514
- PMCID: PMC1287799
- DOI: 10.1128/JCM.43.11.5760-5767.2005
Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003
Abstract
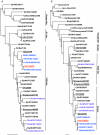
Avian H9N2 influenza A virus has caused repeated human infections in Asia since 1998. Here we report that an H9N2 influenza virus infected a 5-year-old child in Hong Kong in 2003. To identify the possible source of the infection, the human isolate and other H9N2 influenza viruses isolated from Hong Kong poultry markets from January to October 2003 were genetically and antigenically characterized. The findings of this study show that the human H9N2 influenza virus, A/Hong Kong/2108/03, is of purely avian origin and is closely related to some viruses circulating in poultry in the markets of Hong Kong. The continued presence of H9N2 influenza viruses in poultry markets in southern China increases the likelihood of avian-to-human interspecies transmission.
Figures



References
-
- Guo, Y., J. Dong, M. Wang, Y. Zhang, J. Guo, and K. Wu. 2001. Characterization of hemagglutinin gene of influenza A virus subtype H9N2. Chinese Med. J. 114:76-79. - PubMed
-
- Guo, Y., J. Li, X. Cheng, M. Wang, Y. Zhou, C. Li, F. Chai, H. Liao, Y. Zhang, J. Guo, L. Huang, and D. Bei. 1999. Discovery of men infected by avian influenza A (H9N2) virus. Chinese J. Exp. Clin. Virol. 13:105-108. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

