Comparison of microbial community compositions of two subglacial environments reveals a possible role for microbes in chemical weathering processes
- PMID: 16269734
- PMCID: PMC1287656
- DOI: 10.1128/AEM.71.11.6986-6997.2005
Comparison of microbial community compositions of two subglacial environments reveals a possible role for microbes in chemical weathering processes
Abstract
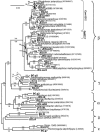
Viable microbes have been detected beneath several geographically distant glaciers underlain by different lithologies, but comparisons of their microbial communities have not previously been made. This study compared the microbial community compositions of samples from two glaciers overlying differing bedrock. Bulk meltwater chemistry indicates that sulfide oxidation and carbonate dissolution account for 90% of the solute flux from Bench Glacier, Alaska, whereas gypsum/anhydrite and carbonate dissolution accounts for the majority of the flux from John Evans Glacier, Ellesmere Island, Nunavut, Canada. The microbial communities were examined using two techniques: clone libraries and dot blot hybridization of 16S rRNA genes. Two hundred twenty-seven clones containing amplified 16S rRNA genes were prepared from subglacial samples, and the gene sequences were analyzed phylogenetically. Although some phylogenetic groups, including the Betaproteobacteria, were abundant in clone libraries from both glaciers, other well-represented groups were found at only one glacier. Group-specific oligonucleotide probes were developed for two phylogenetic clusters that were of particular interest because of their abundance or inferred biochemical capabilities. These probes were used in quantitative dot blot hybridization assays with a range of samples from the two glaciers. In addition to shared phyla at both glaciers, each glacier also harbored a subglacial microbial population that correlated with the observed aqueous geochemistry. These results are consistent with the hypothesis that microbial activity is an important contributor to the solute flux from glaciers.
Figures


References
-
- Anderson, R. S., S. P. Anderson, K. C. MacGregor, E. D. Waddington, S. O'Neel, C. Riihimaki, and M. G. Loso. 2004. Strong feedbacks between hydrology and sliding of a small alpine glacier. J. Geophys. Res. 109:10.1029/2004JF000120.
-
- Anderson, S. P., J. I. Drever, C. D. Frost, and P. Holden. 2000. Chemical weathering in the foreland of a retreating glacier. Geochim. Cosmochim. Acta 64:1173-1189.
-
- Barker, J. D., M. J. Sharp, and R. J. Turner. Organic carbon abundance and dynamics in glacier systems. Submitted for publication.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

