Seasonal variations in virus-host populations in Norwegian coastal waters: focusing on the cyanophage community infecting marine Synechococcus spp
- PMID: 16820451
- PMCID: PMC1489308
- DOI: 10.1128/AEM.00168-06
Seasonal variations in virus-host populations in Norwegian coastal waters: focusing on the cyanophage community infecting marine Synechococcus spp
Abstract
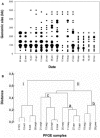
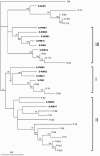
Viruses are ubiquitous components of the marine ecosystem. In the current study we investigated seasonal variations in the viral community in Norwegian coastal waters by pulsed-field gel electrophoresis (PFGE). The results demonstrated that the viral community was diverse, displaying dynamic seasonal variation, and that viral populations of 29 different sizes in the range from 26 to 500 kb were present. Virus populations from 260 to 500 kb and dominating autotrophic pico- and nanoeukaryotes showed similar dynamic variations. Using flow cytometry and real-time PCR, we focused in particular on one host-virus system: Synechococcus spp. and cyanophages. The two groups covaried throughout the year and were found in the highest amounts in fall with concentrations of 7.3 x 10(4) Synechococcus cells ml(-1) and 7.2 x 10(3) cyanophage ml(-1). By using primers targeting the g20 gene in PCRs on DNA extracted from PFGE bands, we demonstrated that cyanophages were found in a genomic size range of 26 to 380 kb. The genetic richness of the cyanophage community, determined by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified g20 gene fragments, revealed seasonal shifts in the populations, with one community dominating in spring and summer and a different one dominating in fall. Phylogenetic analysis of the sequences originating from PFGE and DGGE bands grouped the sequences into three groups, all with homology to cyanomyoviruses present in cultures. Our results show that the cyanophage community in Norwegian coastal waters is dynamic and genetically diverse and has a surprisingly wide genomic size range.
Figures


References
-
- Ackermann, H.-W., and M. S. DuBow. 1987. Viruses of prokaryotes, vol. I. General properties of bacteriophages. CRC Press, Inc., Boca Raton, Fla.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Azam, F., T. Fenchel, T. G. Grey, L. A. Meyer-Reil, and T. Thingstad. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263.
-
- Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

