Genetic diversity of hydrogen-producing bacteria in an acidophilic ethanol-H2-coproducing system, analyzed using the [Fe]-hydrogenase gene
- PMID: 18156331
- PMCID: PMC2258583
- DOI: 10.1128/AEM.01946-07
Genetic diversity of hydrogen-producing bacteria in an acidophilic ethanol-H2-coproducing system, analyzed using the [Fe]-hydrogenase gene
Abstract
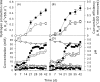
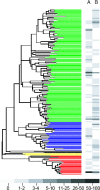
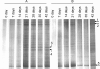
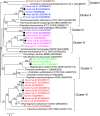
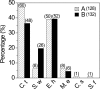
Hydrogen gas (H2) produced by bacterial fermentation of biomass can be a sustainable energy source. The ability to produce H2 gas during anaerobic fermentation was previously thought to be restricted to a few species within the genera Clostridium and Enterobacter. This work reports genomic evidence for the presence of novel H2-producing bacteria (HPB) in acidophilic ethanol-H2-coproducing communities that were enriched using molasses wastewater. The majority of the enriched dominant populations in the acidophilic ethanol-H2-coproducing system were affiliated with low-G+C-content gram-positive bacteria, Bacteroidetes, and Actinobacteria, based on the 16S rRNA gene. However, PCR primers designed to specifically target bacterial hydA yielded 17 unique hydA sequences whose amino acid sequences differed from those of known HPB. The putative ethanol-H2-coproducing bacteria comprised 11 novel phylotypes closely related to Ethanoligenens harbinense, Clostridium thermocellum, and Clostridium saccharoperbutylacetonicum. Furthermore, analysis of the alcohol dehydrogenase isoenzyme also pointed to an E. harbinense-like organism, which is known to have a high conversion rate of carbohydrate to H2 and ethanol. We also found six novel HPB that were associated with lactate-, propionate-, and butyrate-oxidizing bacteria in the acidophilic H2-producing sludge. Thus, the microbial ecology of mesophilic and acidophilic H2 fermentation involves many other bacteria in addition to Clostridium and Enterobacter.
Figures




References
-
- Adams, M. W. W., and E. I. Stiefel. 1998. Biological hydrogen production: not so elementary. Science 282:1842-1843. - PubMed
-
- Ahn, Y., E. J. Park, Y. K. Oh, S. Park, G. Webster, and A. J. Weightman. 2005. Biofilm microbial community of a thermophilic trickling biofilter used for continuous biohydrogen production. FEMS Microbiol. Lett. 249:31-38. - PubMed
-
- Bassam, B. J., G. Caetano-Anolles, and P. M. Gresshoff. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196:80-83. - PubMed
-
- Benemann, J. 1996. Hydrogen biotechnology: progress and prospects. Nat. Biotechnol. 14:1101-1103. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

