A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination
- PMID: 17033965
- PMCID: PMC1698570
- DOI: 10.1086/508709
A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination
Abstract
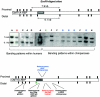
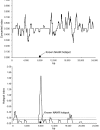
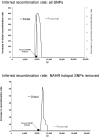
Insights into the origins of structural variation and the mutational mechanisms underlying genomic disorders would be greatly improved by a genomewide map of hotspots of nonallelic homologous recombination (NAHR). Moreover, our understanding of sequence variation within the duplicated sequences that are substrates for NAHR lags far behind that of sequence variation within the single-copy portion of the genome. Perhaps the best-characterized NAHR hotspot lies within the 24-kb-long Charcot-Marie-Tooth disease type 1A (CMT1A)-repeats (REPs) that sponsor deletions and duplications that cause peripheral neuropathies. We investigated structural and sequence diversity within the CMT1A-REPs, both within and between species. We discovered a high frequency of retroelement insertions, accelerated sequence evolution after duplication, extensive paralogous gene conversion, and a greater than twofold enrichment of SNPs in humans relative to the genome average. We identified an allelic recombination hotspot underlying the known NAHR hotspot, which suggests that the two processes are intimately related. Finally, we used our data to develop a novel method for inferring the location of an NAHR hotspot from sequence variation within segmental duplications and applied it to identify a putative NAHR hotspot within the LCR22 repeats that sponsor velocardiofacial syndrome deletions. We propose that a large-scale project to map sequence variation within segmental duplications would reveal a wealth of novel chromosomal-rearrangement hotspots.
Figures







References
Web Resources
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CMT1A-REP sequences [accession numbers DQ480370–DQ480419])
-
- HapMap, http://www.hapmap.org/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CMT1A, HNPP, SMS, NF1, Sotos syndrome, and VCFS)
-
- RepeatMasker, http://www.repeatmasker.org/
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

