Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers
- PMID: 17098925
- PMCID: PMC1796976
- DOI: 10.1128/AEM.01875-06
Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers
Abstract
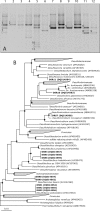
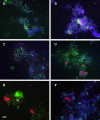
Here we describe the diversity and activity of sulfate-reducing bacteria (SRB) in sulfidogenic bioreactors by using the simultaneous analysis of PCR products obtained from DNA and RNA of the 16S rRNA and dissimilatory sulfite reductase (dsrAB) genes. We subsequently analyzed the amplified gene fragments by using denaturing gradient gel electrophoresis (DGGE). We observed fewer bands in the RNA-based DGGE profiles than in the DNA-based profiles, indicating marked differences in the populations present and in those that were metabolically active at the time of sampling. Comparative sequence analyses of the bands obtained from rRNA and dsrB DGGE profiles were congruent, revealing the same SRB populations. Bioreactors that received either ethanol or isopropanol as an energy source showed the presence of SRB affiliated with Desulfobulbus rhabdoformis and/or Desulfovibrio sulfodismutans, as well as SRB related to the acetate-oxidizing Desulfobacca acetoxidans. The reactor that received wastewater containing a diverse mixture of organic compounds showed the presence of nutritionally versatile SRB affiliated with Desulfosarcina variabilis and another acetate-oxidizing SRB, affiliated with Desulfoarculus baarsii. In addition to DGGE analysis, we performed whole-cell hybridization with fluorescently labeled oligonucleotide probes to estimate the relative abundances of the dominant sulfate-reducing bacterial populations. Desulfobacca acetoxidans-like populations were most dominant (50 to 60%) relative to the total SRB communities, followed by Desulfovibrio-like populations (30 to 40%), and Desulfobulbus-like populations (15 to 20%). This study is the first to identify metabolically active SRB in sulfidogenic bioreactors by using the functional gene dsrAB as a molecular marker. The same approach can also be used to infer the ecological role of coexisting SRB in other habitats.
Figures


References
-
- Bak, F., and N. Pfennig. 1987. Chemolithotrophic growth of Desulfovibrio sulfodismutans sp. nov. by disproportionation of inorganic sulfur compounds. Arch. Microbiol. 147:184-189.
-
- Chang, Y. J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

