Multilocus sequence typing system for the endosymbiont Wolbachia pipientis
- PMID: 16936055
- PMCID: PMC1636189
- DOI: 10.1128/AEM.00731-06
Multilocus sequence typing system for the endosymbiont Wolbachia pipientis
Abstract
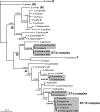
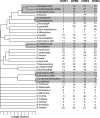
The eubacterial genus Wolbachia comprises one of the most abundant groups of obligate intracellular bacteria, and it has a host range that spans the phyla Arthropoda and Nematoda. Here we developed a multilocus sequence typing (MLST) scheme as a universal genotyping tool for Wolbachia. Internal fragments of five ubiquitous genes (gatB, coxA, hcpA, fbpA, and ftsZ) were chosen, and primers that amplified across the major Wolbachia supergroups found in arthropods, as well as other divergent lineages, were designed. A supplemental typing system using the hypervariable regions of the Wolbachia surface protein (WSP) was also developed. Thirty-seven strains belonging to supergroups A, B, D, and F obtained from singly infected hosts were characterized by using MLST and WSP. The number of alleles per MLST locus ranged from 25 to 31, and the average levels of genetic diversity among alleles were 6.5% to 9.2%. A total of 35 unique allelic profiles were found. The results confirmed that there is a high level of recombination in chromosomal genes. MLST was shown to be effective for detecting diversity among strains within a single host species, as well as for identifying closely related strains found in different arthropod hosts. Identical or similar allelic profiles were obtained for strains harbored by different insect species and causing distinct reproductive phenotypes. Strains with similar WSP sequences can have very different MLST allelic profiles and vice versa, indicating the importance of the MLST approach for strain identification. The MLST system provides a universal and unambiguous tool for strain typing, population genetics, and molecular evolutionary studies. The central database for storing and organizing Wolbachia bacterial and host information can be accessed at http://pubmlst.org/wolbachia/.
Figures




References
-
- Baldo, L., J. D. Bartos, J. H. Werren, C. Bazzocchi, M. Casiraghi, and S. Panelli. 2002. Different rates of nucleotide substitutions in Wolbachia endosymbionts of arthropods and nematodes: arms race or host shifts? Parasitologia 44:179-187. - PubMed
-
- Baldo, L., S. Bordenstein, J. J. Wernegreen, and J. H. Werren. 2006. Widespread recombination throughout Wolbachia genomes. Mol. Biol. Evol. 23:437-449. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

