Integrated genomic characterization of oesophageal carcinoma
- PMID: 28052061
- PMCID: PMC5651175
- DOI: 10.1038/nature20805
Integrated genomic characterization of oesophageal carcinoma
Abstract
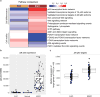
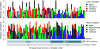
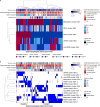
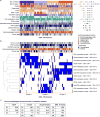
Oesophageal cancers are prominent worldwide; however, there are few targeted therapies and survival rates for these cancers remain dismal. Here we performed a comprehensive molecular analysis of 164 carcinomas of the oesophagus derived from Western and Eastern populations. Beyond known histopathological and epidemiologic distinctions, molecular features differentiated oesophageal squamous cell carcinomas from oesophageal adenocarcinomas. Oesophageal squamous cell carcinomas resembled squamous carcinomas of other organs more than they did oesophageal adenocarcinomas. Our analyses identified three molecular subclasses of oesophageal squamous cell carcinomas, but none showed evidence for an aetiological role of human papillomavirus. Squamous cell carcinomas showed frequent genomic amplifications of CCND1 and SOX2 and/or TP63, whereas ERBB2, VEGFA and GATA4 and GATA6 were more commonly amplified in adenocarcinomas. Oesophageal adenocarcinomas strongly resembled the chromosomally unstable variant of gastric adenocarcinoma, suggesting that these cancers could be considered a single disease entity. However, some molecular features, including DNA hypermethylation, occurred disproportionally in oesophageal adenocarcinomas. These data provide a framework to facilitate more rational categorization of these tumours and a foundation for new therapies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures











Comment in
-
Cancer genomics: Spot the difference.Nature. 2017 Jan 12;541(7636):162-163. doi: 10.1038/nature21112. Epub 2017 Jan 4. Nature. 2017. PMID: 28052059 No abstract available.
-
Not All Esophageal Tumors Equal.Cancer Discov. 2017 Mar;7(3):238. doi: 10.1158/2159-8290.CD-NB2017-006. Epub 2017 Jan 19. Cancer Discov. 2017. PMID: 28104797
-
Genetics: Oesophageal cancer - not all alike.Nat Rev Clin Oncol. 2017 Mar;14(3):138. doi: 10.1038/nrclinonc.2017.9. Epub 2017 Jan 24. Nat Rev Clin Oncol. 2017. PMID: 28117415 No abstract available.
References
-
- De Angelis R, et al. Cancer survival in Europe 1999–2007 by country and age: results of EUROCARE—5-a population-based study. Lancet Oncol. 15:23–34. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. - PubMed
-
- Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 65:87–108. - PubMed
-
- Siewert JR, Ott K. Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat Oncol. 2007;17:38–44. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
- 5U24CA143883/NH/NIH HHS/United States
- U24 CA143866/CA/NCI NIH HHS/United States
- 5U24CA143858/NH/NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- 5U24CA143866/NH/NIH HHS/United States
- U24 CA143840/CA/NCI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U24 CA126543/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U24 CA210949/CA/NCI NIH HHS/United States
- 5U24CA143867/NH/NIH HHS/United States
- 5U24CA143845/NH/NIH HHS/United States
- U24 CA126544/CA/NCI NIH HHS/United States
- UL1 TR000371/TR/NCATS NIH HHS/United States
- 5U24CA143882/NH/NIH HHS/United States
- 5U24CA143835/NH/NIH HHS/United States
- 5U24CA143799/NH/NIH HHS/United States
- U24 CA211000/CA/NCI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- 5U24CA143840/NH/NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143835/CA/NCI NIH HHS/United States
- U24 CA210950/CA/NCI NIH HHS/United States
- 5U24CA143843/NH/NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- U24 CA144025/CA/NCI NIH HHS/United States
- 5U24CA143848/NH/NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U24 CA143843/CA/NCI NIH HHS/United States
- T32 CA009676/CA/NCI NIH HHS/United States
- U24 CA210969/CA/NCI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
- U24 CA210999/CA/NCI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- U24 CA199461/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

