Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence
- PMID: 17416733
- PMCID: PMC1913755
- DOI: 10.1105/tpc.106.042911
Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence
Abstract
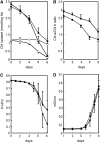
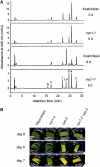
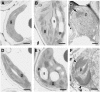
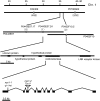
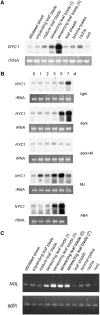
Chlorophyll degradation is an aspect of leaf senescence, which is an active process to salvage nutrients from old tissues. non-yellow coloring1 (nyc1) is a rice (Oryza sativa) stay-green mutant in which chlorophyll degradation during senescence is impaired. Pigment analysis revealed that degradation of not only chlorophylls but also light-harvesting complex II (LHCII)-bound carotenoids was repressed in nyc1, in which most LHCII isoforms were selectively retained during senescence. Ultrastructural analysis of nyc1 chloroplasts revealed that large and thick grana were present even in the late stage of senescence, suggesting that degradation of LHCII is required for the proper degeneration of thylakoid membranes. Map-based cloning of NYC1 revealed that it encodes a chloroplast-localized short-chain dehydrogenase/reductase (SDR) with three transmembrane domains. The predicted structure of the NYC1 protein and the phenotype of the nyc1 mutant suggest the possibility that NYC1 is a chlorophyll b reductase. Although we were unable to detect the chlorophyll b reductase activity of NYC1, NOL (for NYC1-like), a protein closely related to NYC1 in rice, showed chlorophyll b reductase activity in vitro. We suggest that NYC1 and NOL encode chlorophyll b reductases with divergent functions. Our data collectively suggest that the identified SDR protein NYC1 plays essential roles in the regulation of LHCII and thylakoid membrane degradation during senescence.
Figures



References
-
- Allen, J.F., and Forsberg, J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6 317–326. - PubMed
-
- Arimura, S., Takusagawa, S., Hatano, S., Nakazono, M., Hirai, A., and Tsutsumi, N. (1999). A novel plant nuclear gene encoding chloroplast ribosomal protein S9 has a transit peptide related to that of rice chloroplast ribosomal protein L12. FEBS Lett. 450 231–234. - PubMed
-
- Bellemare, G., Bartlett, S.G., and Chua, N.H. (1982). Biosynthesis of chlorophyll a/b-binding polypeptides in wild type and the chlorina-f2 mutant of barley. J. Biol. Chem. 257 7762–7767. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

