Dual selection pressure by drugs and HLA class I-restricted immune responses on human immunodeficiency virus type 1 protease
- PMID: 17202219
- PMCID: PMC1866003
- DOI: 10.1128/JVI.01547-06
Dual selection pressure by drugs and HLA class I-restricted immune responses on human immunodeficiency virus type 1 protease
Abstract
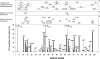
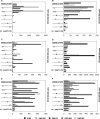
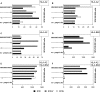
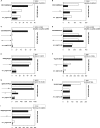
To determine the influence of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cells on the development of drug resistance mutations in the HIV-1 protease, we analyzed protease sequences from viruses from a human leukocyte antigen class I (HLA class I)-typed cohort of 94 HIV-1-positive individuals. In univariate statistical analyses (Fisher's exact test), minor and major drug resistance mutations as well as drug-associated polymorphisms showed associations with HLA class I alleles. All correlations with P values of 0.05 or less were considered to be relevant without corrections for multiple tests. A subset of these observed correlations was experimentally validated by enzyme-linked immunospot assays, allowing the definition of 10 new epitopes recognized by CD8+ T cells from patients with the appropriate HLA class I type. Several drug resistance-associated mutations in the protease acted as escape mutations; however, cells from many patients were still able to generate CD8+ T cells targeting the escape mutants. This result presumably indicates the usage of different T-cell receptors by CD8+ T cells targeting these epitopes in these patients. Our results support a fundamental role for HLA class I-restricted immune responses in shaping the sequence of the HIV-1 protease in vivo. This role may have important clinical implications both for the understanding of drug resistance pathways and for the design of therapeutic vaccines targeting drug-resistant HIV-1.
Figures
References
-
- Alexander, C. S., W. Dong, K. Chan, N. Jahnke, M. V. O'Shaughnessy, T. Mo, M. A. Piaseczny, J. S. Montaner, and P. R. Harrigan. 2001. HIV protease and reverse transcriptase variation and therapy outcome in antiretroviral-naive individuals from a large North American cohort. AIDS 15:601-607. - PubMed
-
- Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H. S. Hewitt, L. Henry, C. H. Linde, J. V. Chisholm III, T. M. Zaman, E. Pae, S. Mallal, B. D. Walker, A. Sette, B. T. Korber, D. Heckerman, and C. Brander. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094-4101. - PubMed
-
- Boehringer Ingelheim Pharmaceuticals. 19April2005, posting date. Tipranavir Anti-Viral Drugs Advisory Committee (AVDAC) briefing document. Boehringer Ingelheim Pharmaceuticals, Ingelheim, Germany. http://www.fda.gov/ohrms/dockets/ac/05/briefing/2005-4139b1-02-boehringe....
-
- Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

