Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra
- PMID: 17664430
- PMCID: PMC1934924
- DOI: 10.1073/pnas.0703375104
Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra
Abstract
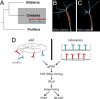
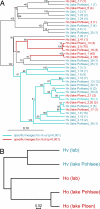
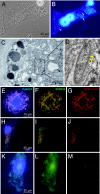
Epithelia in animals are colonized by complex communities of microbes. Although a topic of long-standing interest, understanding the evolution of the microbial communities and their role in triggering innate immune responses has resisted analysis. Cnidaria are among the simplest animals at the tissue grade of organization. To obtain a better understanding of the microbiota associated with phylogenetically ancient epithelia, we have identified the epibiotic and endosymbiotic bacteria of two species of the cnidarian Hydra on the basis of rRNA comparisons. We analyzed individuals of Hydra oligactis and Hydra vulgaris from both laboratory cultures and the wild. We discovered that individuals from both species differ greatly in their bacterial microbiota. Although H. vulgaris polyps have a quite diverse microbiota, H. oligactis appears to be associated with only a limited number of microbes; some of them were found, unexpectedly, to be endosymbionts. Surprisingly, the microfauna showed similar characteristics in individuals of cultures maintained in the laboratory for >30 years and polyps directly isolated from the wild. The significant differences in the microbial communities between the two species and the maintenance of specific microbial communities over long periods of time strongly indicate distinct selective pressures imposed on and within the epithelium. Our analysis suggests that the Hydra epithelium actively selects and shapes its microbial community.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

