Analysis of Euglena gracilis plastid-targeted proteins reveals different classes of transit sequences
- PMID: 16998072
- PMCID: PMC1694827
- DOI: 10.1128/EC.00222-06
Analysis of Euglena gracilis plastid-targeted proteins reveals different classes of transit sequences
Abstract
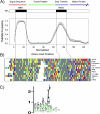
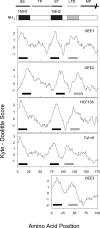
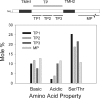
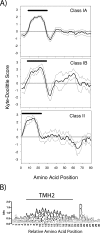
The plastid of Euglena gracilis was acquired secondarily through an endosymbiotic event with a eukaryotic green alga, and as a result, it is surrounded by a third membrane. This membrane complexity raises the question of how the plastid proteins are targeted to and imported into the organelle. To further explore plastid protein targeting in Euglena, we screened a total of 9,461 expressed sequence tag (EST) clusters (derived from 19,013 individual ESTs) for full-length proteins that are plastid localized to characterize their targeting sequences and to infer potential modes of translocation. Of the 117 proteins identified as being potentially plastid localized whose N-terminal targeting sequences could be inferred, 83 were unique and could be classified into two major groups. Class I proteins have tripartite targeting sequences, comprising (in order) an N-terminal signal sequence, a plastid transit peptide domain, and a predicted stop-transfer sequence. Within this class of proteins are the lumen-targeted proteins (class IB), which have an additional hydrophobic domain similar to a signal sequence and required for further targeting across the thylakoid membrane. Class II proteins lack the putative stop-transfer sequence and possess only a signal sequence at the N terminus, followed by what, in amino acid composition, resembles a plastid transit peptide. Unexpectedly, a few unrelated plastid-targeted proteins exhibit highly similar transit sequences, implying either a recent swapping of these domains or a conserved function. This work represents the most comprehensive description to date of transit peptides in Euglena and hints at the complex routes of plastid targeting that must exist in this organism.
Figures



References
-
- Apt, K. E., D. Bhaya, and A. R. Grossman. 1994. Characterization of genes encoding the light-harvesting proteins in diatoms: biogenesis of the fucoxanthin chlorophyll a/c protein complex. J. Appl. Phycol. 6:225-230.
-
- Apt, K. E., N. E. Hoffman, and A. R. Grossman. 1993. The γ-subunit of R-phycoerythrin and its possible mode of transport into the plastid of red algae. J. Biol. Chem. 268:16208-16215. - PubMed
-
- Apt, K. E., L. Zaslavkaia, J. C. Lippmeier, M. Lang, O. Kilian, R. Wetherbee, A. R. Grossman, and P. G. Kroth. 2002. In vivo characterization of diatom multipartite plastid targeting signals. J. Cell Sci. 115:4061-4069. - PubMed
-
- Arimura, S.-I., S. Takusagawa, S. Hatano, M. Nakazono, A. Hirai, and N. Tsutsumi. 1999. A novel plant nuclear gene encoding chloroplast ribosomal protein S9 has a transit peptide related to that of rice chloroplast ribosomal protein L12. FEBS Lett. 450:231-234. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

