Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread
- PMID: 31358985
- PMCID: PMC7244338
- DOI: 10.1038/s41564-019-0492-8
Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread
Abstract
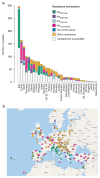
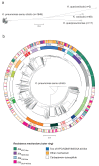
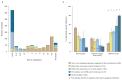
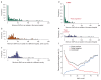
Public health interventions to control the current epidemic of carbapenem-resistant Klebsiella pneumoniae rely on a comprehensive understanding of its emergence and spread over a wide range of geographical scales. We analysed the genome sequences and epidemiological data of >1,700 K. pneumoniae samples isolated from patients in 244 hospitals in 32 countries during the European Survey of Carbapenemase-Producing Enterobacteriaceae. We demonstrate that carbapenemase acquisition is the main cause of carbapenem resistance and that it occurred across diverse phylogenetic backgrounds. However, 477 of 682 (69.9%) carbapenemase-positive isolates are concentrated in four clonal lineages, sequence types 11, 15, 101, 258/512 and their derivatives. Combined analysis of the genetic and geographic distances between isolates with different β-lactam resistance determinants suggests that the propensity of K. pneumoniae to spread in hospital environments correlates with the degree of resistance and that carbapenemase-positive isolates have the highest transmissibility. Indeed, we found that over half of the hospitals that contributed carbapenemase-positive isolates probably experienced within-hospital transmission, and interhospital spread is far more frequent within, rather than between, countries. Finally, we propose a value of 21 for the number of single nucleotide polymorphisms that optimizes the discrimination of hospital clusters and detail the international spread of the successful epidemic lineage, ST258/512.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Genomic epidemiology of carbapenem-resistant Enterobacterales at a New York City hospital over a 10-year period reveals complex plasmid-clone dynamics and evidence for frequent horizontal transfer of bla KPC.Genome Res. 2024 Nov 20;34(11):1895-1907. doi: 10.1101/gr.279355.124. Genome Res. 2024. PMID: 39366703 Free PMC article.
-
Molecular characterization of carbapenem-resistant Enterobacteriaceae and emergence of tigecycline non-susceptible strains in the Henan province in China: a multicentrer study.J Med Microbiol. 2021 Mar;70(3):001325. doi: 10.1099/jmm.0.001325. Epub 2021 Feb 15. J Med Microbiol. 2021. PMID: 33587030 Free PMC article.
-
Genomic and clinical characterisation of multidrug-resistant carbapenemase-producing ST231 and ST16 Klebsiella pneumoniae isolates colonising patients at Siriraj hospital, Bangkok, Thailand from 2015 to 2017.BMC Infect Dis. 2021 Feb 4;21(1):142. doi: 10.1186/s12879-021-05790-9. BMC Infect Dis. 2021. PMID: 33541274 Free PMC article.
-
Systematic review of carbapenem-resistant Enterobacteriaceae causing neonatal sepsis in China.Ann Clin Microbiol Antimicrob. 2019 Nov 14;18(1):36. doi: 10.1186/s12941-019-0334-9. Ann Clin Microbiol Antimicrob. 2019. PMID: 31727088 Free PMC article.
-
[Carbapenemase producing Enterobacteriaceae in the Netherlands: unnoticed spread to several regions].Ned Tijdschr Geneeskd. 2017;161:D1585. Ned Tijdschr Geneeskd. 2017. PMID: 29076442 Review. Dutch.
Cited by
-
Necroptosis in Pneumonia: Therapeutic Strategies and Future Perspectives.Viruses. 2024 Jan 7;16(1):94. doi: 10.3390/v16010094. Viruses. 2024. PMID: 38257794 Free PMC article. Review.
-
A large-scale genomic snapshot of Klebsiella spp. isolates in Northern Italy reveals limited transmission between clinical and non-clinical settings.Nat Microbiol. 2022 Dec;7(12):2054-2067. doi: 10.1038/s41564-022-01263-0. Epub 2022 Nov 21. Nat Microbiol. 2022. PMID: 36411354 Free PMC article.
-
Antimicrobial resistance level and conjugation permissiveness shape plasmid distribution in clinical enterobacteria.Proc Natl Acad Sci U S A. 2023 Dec 19;120(51):e2314135120. doi: 10.1073/pnas.2314135120. Epub 2023 Dec 14. Proc Natl Acad Sci U S A. 2023. PMID: 38096417 Free PMC article.
-
Transformation of microbiology data into a standardised data representation using OpenEHR.Sci Rep. 2021 May 18;11(1):10556. doi: 10.1038/s41598-021-89796-y. Sci Rep. 2021. PMID: 34006956 Free PMC article.
-
Transmission Dynamics and Novel Treatments of High Risk Carbapenem-Resistant Klebsiella pneumoniae: The Lens of One Health.Pharmaceuticals (Basel). 2024 Sep 12;17(9):1206. doi: 10.3390/ph17091206. Pharmaceuticals (Basel). 2024. PMID: 39338368 Free PMC article. Review.
References
-
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. World Health Organisation; 2017.
-
- Cassini A, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infectious Diseases. 2019;19:56–66. doi: 10.1016/S1473-3099(18)306054. - DOI - PMC - PubMed
-
- Martin J, et al. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. Journal of Antimicrobial Chemotherapy. 2017;72:3025–3034. doi: 10.1093/jac/dkx264. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

