Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume
- PMID: 18083871
- PMCID: PMC2227732
- DOI: 10.1128/AEM.01951-07
Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume
Abstract
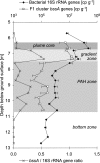
Microbial degradation is the only sustainable component of natural attenuation in contaminated groundwater environments, yet its controls, especially in anaerobic aquifers, are still poorly understood. Hence, putative spatial correlations between specific populations of key microbial players and the occurrence of respective degradation processes remain to be unraveled. We therefore characterized microbial community distribution across a high-resolution depth profile of a tar oil-impacted aquifer where benzene, toluene, ethylbenzene, and xylene (BTEX) degradation depends mainly on sulfate reduction. We conducted depth-resolved terminal restriction fragment length polymorphism fingerprinting and quantitative PCR of bacterial 16S rRNA and benzylsuccinate synthase genes (bssA) to quantify the distribution of total microbiota and specific anaerobic toluene degraders. We show that a highly specialized degrader community of microbes related to known deltaproteobacterial iron and sulfate reducers (Geobacter and Desulfocapsa spp.), as well as clostridial fermenters (Sedimentibacter spp.), resides within the biogeochemical gradient zone underneath the highly contaminated plume core. This zone, where BTEX compounds and sulfate--an important electron acceptor--meet, also harbors a surprisingly high abundance of the yet-unidentified anaerobic toluene degraders carrying the previously detected F1-cluster bssA genes (C. Winderl, S. Schaefer, and T. Lueders, Environ. Microbiol. 9:1035-1046, 2007). Our data suggest that this biogeochemical gradient zone is a hot spot of anaerobic toluene degradation. These findings show that the distribution of specific aquifer microbiota and degradation processes in contaminated aquifers are tightly coupled, which may be of value for the assessment and prediction of natural attenuation based on intrinsic aquifer microbiota.
Figures



References
-
- Anderson, R. T., and D. R. Lovley. 1997. Ecology and biogeochemistry of in situ groundwater bioremediation. Adv. Microb. Ecol. 15:289-350.
-
- Anneser, B., F. Einsiedl, R. U. Meckenstock, L. Richters, F. Wisotzky, and C. Griebler. High-resolution monitoring of biogeochemical gradients in a tar-oil contaminated aquifer. Appl. Geochem., in press.
-
- Bauer, R. D., Y. Zhang, P. Maloszewski, R. U. Meckenstock, and C. Griebler. Aerobic and anaerobic degradation in anoxic toluene plumes—2D microcosm studies. J. Contam. Hydrol., in press. - PubMed
-
- Bekins, B. A., I. M. Cozzarelli, E. M. Godsy, E. Warren, H. I. Essaid, and M. E. Tuccillo. 2001. Progression of natural attenuation processes at a crude oil spill site. II. Controls on spatial distribution of microbial populations. J. Contam. Hydrol. 53:387-406. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

