Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: A comparative analysis of the protocadherin cluster
- PMID: 18319338
- PMCID: PMC2268768
- DOI: 10.1073/pnas.0800398105
Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: A comparative analysis of the protocadherin cluster
Abstract
Cartilaginous fishes are the oldest living phylogenetic group of jawed vertebrates. Here, we demonstrate the value of cartilaginous fish sequences in reconstructing the evolutionary history of vertebrate genomes by sequencing the protocadherin cluster in the relatively small genome (910 Mb) of the elephant shark (Callorhinchus milii). Human and coelacanth contain a single protocadherin cluster with 53 and 49 genes, respectively, that are organized in three subclusters, Pcdhalpha, Pcdhbeta, and Pcdhgamma, whereas the duplicated protocadherin clusters in fugu and zebrafish contain >77 and 107 genes, respectively, that are organized in Pcdhalpha and Pcdhgamma subclusters. By contrast, the elephant shark contains a single protocadherin cluster with 47 genes organized in four subclusters (Pcdhdelta, Pcdhepsilon, Pcdhmu, and Pcdhnu). By comparison with elephant shark sequences, we discovered a Pcdhdelta subcluster in teleost fishes, coelacanth, Xenopus, and chicken. Our results suggest that the protocadherin cluster in the ancestral jawed vertebrate contained more subclusters than modern vertebrates, and the evolution of the protocadherin cluster is characterized by lineage-specific differential loss of entire subclusters of genes. In contrast to teleost fish and mammalian protocadherin genes that have undergone gene conversion events, elephant shark protocadherin genes have experienced very little gene conversion. The syntenic block of genes in the elephant shark protocadherin locus is well conserved in human but disrupted in fugu. Thus, the elephant shark genome appears to be less prone to rearrangements compared with teleost fish genomes. The small and "stable" genome of the elephant shark is a valuable reference for understanding the evolution of vertebrate genomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
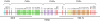


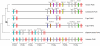
References
-
- Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

