The Foldback-like element Galileo belongs to the P superfamily of DNA transposons and is widespread within the Drosophila genus
- PMID: 18287066
- PMCID: PMC2268567
- DOI: 10.1073/pnas.0712110105
The Foldback-like element Galileo belongs to the P superfamily of DNA transposons and is widespread within the Drosophila genus
Abstract
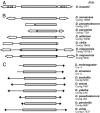
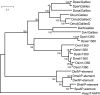
Galileo is the only transposable element (TE) known to have generated natural chromosomal inversions in the genus Drosophila. It was discovered in Drosophila buzzatii and classified as a Foldback-like element because of its long, internally repetitive, terminal inverted repeats (TIRs) and lack of coding capacity. Here, we characterized a seemingly complete copy of Galileo from the D. buzzatii genome. It is 5,406 bp long, possesses 1,229-bp TIRs, and encodes a 912-aa transposase similar to those of the Drosophila melanogaster 1360 (Hoppel) and P elements. We also searched the recently available genome sequences of 12 Drosophila species for elements similar to Dbuz\Galileo by using bioinformatic tools. Galileo was found in six species (ananassae, willistoni, peudoobscura, persimilis, virilis, and mojavensis) from the two main lineages within the Drosophila genus. Our observations place Galileo within the P superfamily of cut-and-paste transposons and extend considerably its phylogenetic distribution. The interspecific distribution of Galileo indicates an ancient presence in the genus, but the phylogenetic tree built with the transposase amino acid sequences contrasts significantly with that of the species, indicating lineage sorting and/or horizontal transfer events. Our results also suggest that Foldback-like elements such as Galileo may evolve from DNA-based transposon ancestors by loss of the transposase gene and disproportionate elongation of TIRs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kidwell MG, Lisch DR. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: American Society for Microbiology; 2002. pp. 59–89.
-
- Capy P, Bazin C, Higuet D, Langin T. Dynamics and Evolution of Transposable Elements. Heidelberg: Springer; 1998.
-
- Haren L, Ton-Hoang B, Chandler M. Integrating DNA: transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. - PubMed
-
- Rio DC. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: American Society for Microbiology; 2002. pp. 484–518.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

